Unlocking the Nano-Scale: How AFM Revolutionizes 3D Printed Biomedical Material Surface Analysis
This article provides a comprehensive guide for researchers and biomedical engineers on applying Atomic Force Microscopy (AFM) to characterize 3D-printed materials.
Unlocking the Nano-Scale: How AFM Revolutionizes 3D Printed Biomedical Material Surface Analysis
Abstract
This article provides a comprehensive guide for researchers and biomedical engineers on applying Atomic Force Microscopy (AFM) to characterize 3D-printed materials. We explore the fundamental principles of AFM, detailing practical methodologies for analyzing surface topography, roughness, and mechanical properties. The guide addresses common troubleshooting scenarios and offers optimization strategies for reliable data acquisition. Finally, we validate AFM's role by comparing it with complementary techniques like SEM and profilometry, concluding with its critical implications for ensuring the quality, functionality, and safety of 3D-printed medical devices, implants, and drug delivery systems.
Why Nano-Scale Surface Analysis is Critical for 3D Printed Biomedical Materials
The Surface-Microstructure-Function Paradigm in 3D Printed Medical Devices
This document provides Application Notes and Protocols for investigating the Surface-Microstructure-Function (SMF) Paradigm in 3D-printed medical devices, framed within a thesis on Atomic Force Microscopy (AFM) for 3D printing material surface analysis. The surface microstructure, dictated by the additive manufacturing process, directly influences critical functional outcomes such as protein adsorption, cellular response, bacterial adhesion, and drug release kinetics.
The SMF paradigm posits a direct causal chain: Printing Parameters → Surface Microstructure → Biological/Functional Response. The following tables summarize key quantitative relationships established in recent literature.
Table 1: Influence of Printing Parameters on Surface Roughness (Sa)
| 3D Printing Technology | Material | Layer Height (µm) | Nozzle Temp (°C) | Measured Sa (nm) | Primary Analysis Technique |
|---|---|---|---|---|---|
| Fused Deposition Modeling (FDM) | PCL | 100 | 110 | 1,520 ± 210 | AFM (10µm scan) |
| Fused Deposition Modeling (FDM) | PCL | 200 | 110 | 3,850 ± 450 | AFM (10µm scan) |
| Stereolithography (SLA) | Resin (Biocompatible) | 25 | N/A | 45 ± 12 | AFM (5µm scan) |
| Selective Laser Sintering (SLS) | PA12 (Nylon) | 80 | N/A | 18 ± 5 | AFM (5µm scan) |
| Direct Ink Writing (DIW) | Alginate-Gelatin | 150 | 22 | 890 ± 140 | Confocal Profilometry |
Table 2: Functional Outcomes Correlated with Surface Microstructure
| Device Application | Material | Surface Roughness (Sa) | Pore Size (µm) | Key Functional Outcome | Measurement |
|---|---|---|---|---|---|
| Bone Scaffold | β-TCP (SLS) | ~22 µm | 350 | Osteoblast adhesion density (Day 7) | 85% coverage |
| Bone Scaffold | β-TCP (SLS) | ~55 µm | 350 | Osteoblast adhesion density (Day 7) | 96% coverage |
| Drug-Eluting Implant | PLA (FDM) | 1.2 µm | N/A | Burst Release (First 24h) | 38% of payload |
| Drug-Eluting Implant | PLA (FDM) | 0.3 µm* | N/A | Burst Release (First 24h) | 12% of payload |
| Antimicrobial Surface | ABS (FDM) | 4.5 µm | N/A | S. aureus adhesion (CFU/cm²) | 1.2 x 10⁵ |
| Antimicrobial Surface | ABS (FDM) | 0.8 µm* | N/A | S. aureus adhesion (CFU/cm²) | 2.7 x 10⁴ |
Note: Post-processing (e.g., solvent vapor smoothing) applied.
Experimental Protocols
Protocol 1: AFM-Based Topographical and Mechanical Mapping of 3D-Printed Surfaces
Objective: To quantitatively characterize the surface roughness, texture, and nanomechanical properties of a 3D-printed medical device sample. Materials: See "Scientist's Toolkit" (Section 6). Procedure:
- Sample Preparation: Cut printed sample to fit AFM sample disk (~1cm x 1cm). Clean with compressed air or inert gas to remove loose particles. For polymers, avoid solvent cleaning unless its effect is under study. Mount securely with double-sided tape or adhesive.
- AFM Calibration: Calibrate the cantilever deflection sensitivity on a clean, rigid surface (e.g., sapphire). Perform thermal tune to determine the spring constant of the cantilever.
- Scanning Parameters:
- Mode: Use PeakForce Tapping or Quantitative Imaging (QI) mode for simultaneous topographical and mechanical property mapping.
- Probe: Use a silicon tip with a nominal spring constant of 0.4-4 N/m and resonant frequency of 150-300 kHz in air.
- Scan Size: 1 µm², 10 µm², and 50 µm² areas to capture features across scales.
- Resolution: 512 x 512 pixels.
- PeakForce Setpoint: Adjust to maintain a consistent, non-destructive force (typically 1-10 nN).
- Data Acquisition: Acquire a minimum of three random, non-overlapping scans per sample condition (n≥3 samples per print parameter set). Record height, PeakForce Error, and DMT Modulus channels.
- Analysis:
- Roughness: Apply a first-order plane fit to height data. Calculate Sa (Average Roughness), Sq (RMS Roughness), and Sz (Maximum Height) per ISO 25178 standards.
- Texture: Use Fast Fourier Transform (FFT) analysis to identify dominant periodicities from layer lines or toolpaths.
- Modulus: Generate histograms of the DMT modulus values from the mapped area, reporting the median and interquartile range.
Protocol 2: In Vitro Protein Adsorption and Cell Adhesion Assay
Objective: To link AFM-characterized surface microstructure to early biological response. Materials: Sample discs, Dulbecco’s Modified Eagle Medium (DMEM), Fetal Bovine Serum (FBS), Fibronectin, PBS, MC3T3-E1 osteoprogenitor cells, Calcein-AM live stain, 4% Paraformaldehyde (PFA), Triton X-100, Phalloidin (actin stain), DAPI. Procedure:
- Surface Characterization: Perform AFM analysis per Protocol 1 on a representative sample from each batch.
- Protein Pre-conditioning: Sterilize samples (70% ethanol, UV irradiation). Immerse in complete cell culture medium (DMEM + 10% FBS) or a solution of 10 µg/mL fibronectin in PBS. Incubate at 37°C for 1 hour.
- Cell Seeding: Seed MC3T3-E1 cells at a density of 10,000 cells/cm² onto samples in 24-well plates. Allow to adhere for 4 hours.
- Fixation and Staining: At adhesion timepoint (e.g., 4h, 24h), rinse with PBS, fix with 4% PFA for 15 min, permeabilize with 0.1% Triton X-100 for 5 min. Stain actin cytoskeleton with Phalloidin (1:500) and nuclei with DAPI (1:1000). Image via confocal microscopy.
- Quantification: Use ImageJ to count nuclei for adhesion density. Analyze cell spread area and cytoskeletal morphology from phalloidin images. Correlate metrics with AFM-derived Sa and Sz values.
Protocol 3: Drug Release Kinetics from Microstructured Surfaces
Objective: To quantify how surface area and texture from printing affect drug elution profile. Materials: Drug-loaded filament (e.g., PLA with 5% w/w Rifampicin), phosphate-buffered saline (PBS, pH 7.4), 0.1% w/v Tween 80 (to maintain sink conditions), UV-Vis Spectrophotometer or HPLC. Procedure:
- Fabrication: Print identical disc geometries (e.g., Ø10mm x 1mm) varying only layer height (e.g., 100 µm vs. 200 µm). Weigh each disc accurately.
- Release Study: Place each disc in a sealed container with 10 mL of release medium (PBS + 0.1% Tween 80) at 37°C under gentle agitation (50 rpm). Maintain sink conditions.
- Sampling: At predetermined timepoints (0.5, 1, 2, 4, 8, 24, 48, 72, 168 h), withdraw 1 mL of medium and replace with fresh pre-warmed medium.
- Analysis: Quantify drug concentration in sampled medium via UV-Vis at λmax (e.g., 475 nm for Rifampicin). Use a standard curve for absolute quantification.
- Modeling: Plot cumulative release (%) vs. time. Fit data to models: Zero-order, First-order, Higuchi (diffusion-controlled), and Korsmeyer-Peppas (to determine release exponent n). Correlate release rate constants with AFM-measured surface area and roughness.
Signaling Pathways & Experimental Workflows
Diagram Title: SMF Paradigm & Cell Signaling Pathway
Diagram Title: Integrated SMF Experimental Workflow
The Scientist's Toolkit: Research Reagent & Material Solutions
Table 3: Essential Materials for SMF Paradigm Research
| Item | Function/Application | Key Considerations |
|---|---|---|
| AFM with PeakForce Tapping/QI Mode | Nanoscale topographic & mechanical mapping. Essential for quantifying Sa, modulus. | Must handle sample roughness up to 10-15µm. Environmental control is beneficial. |
| Silicon AFM Probes (SCANASYST-AIR) | For high-resolution imaging of polymers. Spring constant ~0.4 N/m. | Optimized for PeakForce Tapping. Blunt tip for durability on rough surfaces. |
| Biocompatible 3D Printing Resins (e.g., Dental SG, MED610) | For SLA-printed devices needing cytocompatibility. | Check ISO 10993 certifications. Post-curing affects surface energy. |
| Drug-Loaded Thermoplastic Filaments (e.g., PLA + Antibiotic) | For fabricating drug-eluting study samples via FDM. | Ensure homogeneous drug dispersion. Hot-end temperature critical for stability. |
| Solvent Vapor Smoothing Station (e.g., for ABS) | For post-processing to reduce surface roughness. A key experimental variable. | Use controlled solvent (e.g., acetone) exposure times. Conduct in fume hood. |
| Fibronectin, Fluorescently Conjugated | For quantifying and visualizing protein adsorption onto microstructured surfaces. | Use consistent concentration and incubation time. BSA blocking step required. |
| Calcein-AM / Ethidium Homodimer-1 Live/Dead Assay Kit | For rapid viability assessment of cells on test surfaces. | Optimize dye concentration for porous/microstructured surfaces. |
| Rhodamine-Phalloidin & DAPI | For staining F-actin and nuclei to quantify cell spreading and morphology. | Permeabilization time may vary with material porosity. |
| Simulated Body Fluid (SBF) | For assessing apatite formation (bioactivity) on bone implant surfaces. | Solution must be prepared and used under strict, stable temperature conditions. |
| HPLC System with PDA Detector | For accurate quantification of drug concentrations in release kinetics studies (Protocol 3). | Superior to UV-Vis for complex media or degraded products. |
Principles and Instrumentation
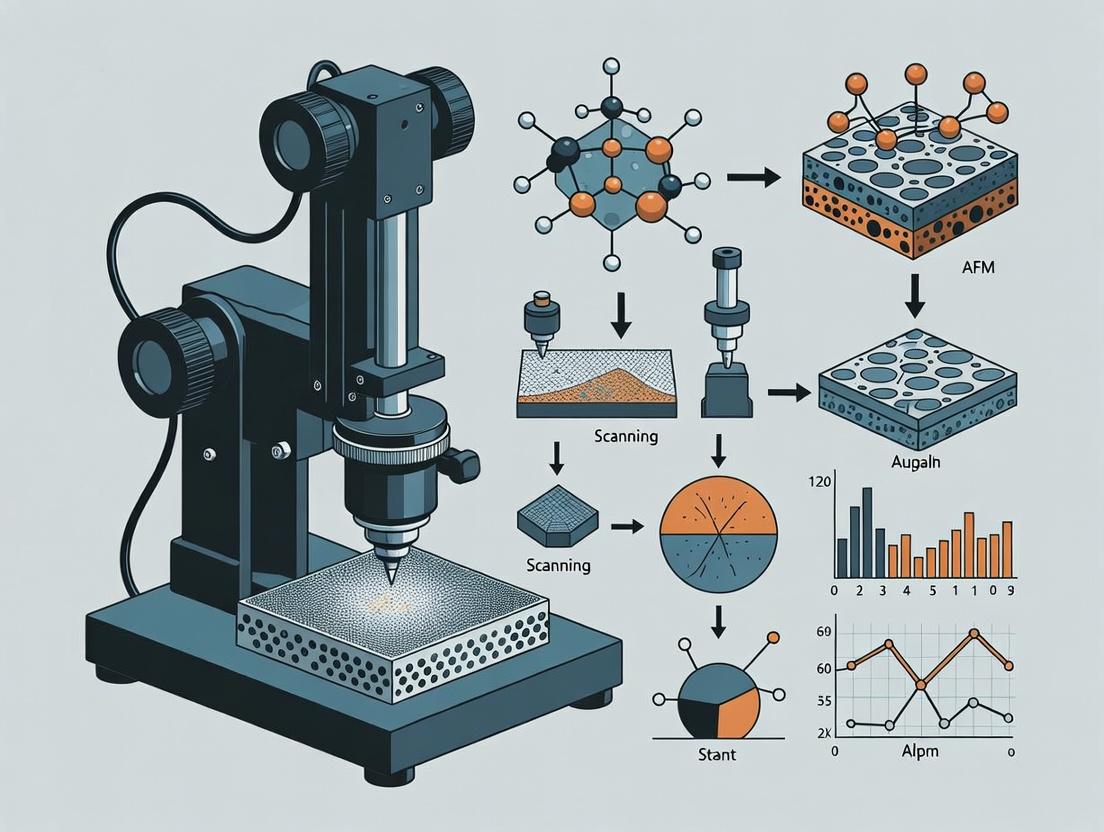
Atomic Force Microscopy (AFM) is a high-resolution scanning probe technique capable of characterizing surface topography and a wide range of physical properties at the nanoscale. Its operation is based on the mechanical interaction between a sharp tip mounted on a flexible cantilever and the sample surface. A laser beam reflected off the back of the cantilever onto a photodetector monitors cantilever deflection, which is used to generate a three-dimensional surface map. For 3D printing material research, AFM is indispensable for quantifying surface roughness, layer adhesion, porosity, and nanomechanical properties, directly correlating print parameters with final material performance.
Table 1: Key Quantitative Parameters in AFM for Material Science
| Parameter | Typical Range | Relevance to 3D-Printed Materials |
|---|---|---|
| Lateral (XY) Resolution | 0.1 - 10 nm | Resolves individual polymer strands, filler particles, and layer boundaries. |
| Vertical (Z) Resolution | 0.01 - 0.1 nm | Measures step heights between printed layers and surface roughness precisely. |
| Force Sensitivity | 1 - 100 pN | Critical for measuring adhesion between layers and local mechanical properties. |
| Scan Size Range | 100 nm - 100+ μm | Enables analysis from nanoscale features to macro-scale print artifacts. |
| Typical Scan Rate | 0.5 - 2 lines/sec | Balances imaging speed with resolution and force control to prevent sample damage. |
Primary Operational Modes
Contact Mode
The tip scans in constant physical contact with the surface. The deflection of the cantilever is kept constant by a feedback loop that adjusts the scanner height. This mode provides high-resolution topographic imaging and frictional force data.
- Advantage: Fast scanning, good for rough, stiff samples.
- Disadvantage: Lateral forces can deform or damage soft materials (e.g., many polymers).
Intermittent Contact (Tapping) Mode
The cantilever is oscillated at or near its resonant frequency. The tip intermittently contacts the surface, and the change in oscillation amplitude or phase is used for feedback. This is the most common mode for 3D-printed polymer analysis.
- Advantage: Minimizes lateral forces, excellent for soft, adhesive, or easily damaged samples.
- Disadvantage: Slightly slower than contact mode.
Non-Contact Mode
The cantilever oscillates near the surface without making contact, sensing van der Waals forces. It is rarely used for polymers due to their common adhesive nature.
- Advantage: Extremely low force, no sample deformation.
- Disadvantage: Lower resolution, requires ultra-clean surfaces in vacuum/air.
Table 2: Comparison of Primary AFM Imaging Modes for Polymer Analysis
| Mode | Feedback Signal | Force Applied | Best For 3D-Printed Materials | Risk of Damage |
|---|---|---|---|---|
| Contact | Cantilever Deflection | High (Constant) | Stiff composites (e.g., carbon-fiber filled), cured resins | High for soft materials |
| Tapping | Oscillation Amplitude | Low (Intermittent) | Most polymers, hydrogels, TPUs, surface roughness | Very Low |
| Non-Contact | Oscillation Frequency/Phase | Very Low | Rare; potentially for delicate, non-adhesive top coatings | Negligible |
Advanced Modes for Material Property Mapping
Force Spectroscopy & Nanoindentation
The AFM tip is used as a nanoindenter. By recording the force-distance curve at a single point or an array of points, local mechanical properties like Young's modulus, adhesion force, and deformation can be quantified.
Protocol: Nanoindentation Mapping of a 3D-Printed Polymer Blend
Objective: To map the spatial variation of elastic modulus across the interface between two co-printed polymers. Materials: See "The Scientist's Toolkit" below. Procedure:
- Sample Preparation: Mount a cross-sectioned sample of the printed interface on a standard AFM specimen disk using a conductive adhesive tab. Ensure the surface is clean and level.
- Cantilever Selection & Calibration: Use a silicon cantilever with a calibrated spring constant (k, typically 1-50 N/m) and a known tip radius (R, via SEM or calibration grating). Determine the optical lever sensitivity (InvOLS) on a clean, rigid sapphire surface.
- Topography Imaging: First, acquire a high-resolution tapping mode image of the target interface region to identify the scan area for the property map.
- Force Curve Array Setup: Define a grid (e.g., 64x64 points) over the region of interest. Set the maximum applied force (setpoint) to 50-200 nN to avoid plastic deformation. Define the extension/retraction speed (typically 0.5-2 μm/s).
- Data Acquisition: Automatically acquire a force-distance curve at every point in the grid.
- Data Analysis: Fit the retraction portion of each curve with an appropriate contact mechanics model (e.g., Derjaguin–Muller–Toporov (DMT) model for stiff samples, or Oliver-Pharr for hard materials). The reduced elastic modulus (E*) is calculated from the slope of the unloading curve. Generate a 2D map of modulus vs. position.
Table 3: Representative Nanoindentation Data from a PLA-TPU Interface
| Position Relative to Interface | Average Reduced Modulus (E*) | Standard Deviation | Adhesion Force |
|---|---|---|---|
| PLA Region (5 μm away) | 3.5 GPa | ± 0.4 GPa | 15 nN |
| Interface (0 μm) | 1.2 GPa | ± 0.6 GPa | 65 nN |
| TPU Region (5 μm away) | 55 MPa | ± 12 MPa | 120 nN |
Phase Imaging (in Tapping Mode)
Monitors the phase lag between the driving oscillation and the cantilever response. This signal is sensitive to viscoelastic properties, adhesion, and dissipation energy, highlighting areas of different material composition.
Conductive AFM (C-AFM)
A conductive tip is used in contact mode with a voltage bias applied. It measures local conductivity or current flow, useful for analyzing printed electronics or composites with conductive fillers (e.g., graphene, CNTs).
Application Workflow for 3D Printing Material Analysis
AFM Workflow for 3D-Printed Material Analysis
The Scientist's Toolkit
Table 4: Essential Research Reagent Solutions & Materials for AFM Analysis of 3D-Printed Polymers
| Item | Function & Relevance |
|---|---|
| Conductive Adhesive Tabs/Carbon Tape | Securely mounts non-magnetic, insulating polymer samples to the AFM metal stub to prevent charging and drift. |
| Pulsed Force Mode (PFM) Cantilevers | Specialized cantilevers with well-defined spring constants and tips for quantitative nanomechanical mapping. |
| Diamond-Coated Tips | For repeated nanoindentation on hard composite materials (e.g., ceramic-filled resins) to maintain tip geometry. |
| Calibration Gratings (TGZ1, PG, HS) | Essential for verifying scanner accuracy in X, Y, and Z, and for tip morphology characterization post-scan. |
| Non-Acoustic Enclosure/Anti-Vibration Table | Isolates the AFM from environmental vibrations crucial for achieving high-resolution data on all length scales. |
| Anti-Static Gun | Neutralizes static charge on polymer samples, which can cause imaging artifacts and attract dust. |
| Soft Polymer Reference Samples (PDMS) | Used to validate force curve calibration and nanoindentation protocols on materials of known, low modulus. |
Within the context of a broader thesis on Atomic Force Microscopy (AFM) for 3D printing material surface analysis, characterizing key surface parameters is fundamental. For 3D-printed biomaterials, especially in drug delivery and tissue engineering, surface topography, roughness, and mechanical properties directly influence cellular adhesion, proliferation, differentiation, and drug release kinetics. This document provides application notes and detailed protocols for the quantitative assessment of these parameters using AFM.
Table 1: Typical Surface Roughness and Mechanical Property Ranges for Common 3D-Printed Biomedical Polymers
| Material & Printing Method | Avg. Roughness, Ra (nm) | RMS Roughness, Rq (nm) | Reduced Elastic Modulus, Er (MPa) | Adhesion Force (nN) | Key Application Context |
|---|---|---|---|---|---|
| PCL (FDM) | 250 - 850 | 300 - 1050 | 120 - 250 | 15 - 40 | Soft tissue scaffolds, drug-eluting implants |
| PLA (FDM) | 150 - 600 | 200 - 750 | 2000 - 3500 | 8 - 25 | Structural scaffolds, orthopedic templates |
| SLA Resin (Standard) | 10 - 50 | 15 - 65 | 1500 - 3000 | 20 - 60 | Microfluidic devices, high-res. prototypes |
| Alginate/Gelatin (Bioprinting) | 50 - 200 | 70 - 250 | 5 - 50 | 40 - 120 | Cell-laden hydrogels, tissue models |
| TPU (FDM) | 300 - 1000 | 400 - 1250 | 30 - 100 | 30 - 80 | Flexible/elastomeric implants |
Table 2: Impact of Key Surface Parameters on Biological Responses in Drug Development Research
| Surface Parameter | Target Range for Enhanced Cell Response | Influence on Drug Release/Pharmacology | Recommended AFM Mode |
|---|---|---|---|
| Ra (Sub-100 nm) | Fibroblast adhesion, endothelialization | Modulates protein adsorption, affecting release kinetics | Tapping Mode, Contact Mode |
| Ra (100-1000 nm) | Osteoblast differentiation, mesenchymal stem cell fate | Increased surface area can accelerate burst release | Tapping Mode |
| Elastic Modulus (1-10 kPa) | Neural progenitor cell differentiation | Affects degradation rate of polymer matrix | Force Spectroscopy (QNM) |
| Elastic Modulus (10-100 kPa) | Muscle cell maturation | Influences mechanical integrity of drug depot | Force Spectroscopy (QNM) |
| Adhesion Force (High) | Platelet adhesion (thrombogenicity) | Can trap carrier particles or proteins | Force Spectroscopy |
| Adhesion Force (Moderate) | Controlled cell spreading and signaling | Optimal for targeted nanoparticle binding | Force Spectroscopy |
Experimental Protocols
Protocol 1: Topography and Roughness (Ra, Rq) Mapping of 3D-Printed Surfaces
Objective: To acquire high-resolution 3D topography and calculate ISO 4287-compliant roughness parameters.
Materials & Sample Prep:
- Sample: 3D-printed polymer scaffold (e.g., PCL, PLA). Solvent-clean if necessary (e.g., ethanol wash, air dry).
- AFM Probe: Silicon nitride tip (e.g., Bruker SNL, k ≈ 0.35 N/m) for contact mode, or silicon tip (e.g., Tap150Al-G, f₀ ≈ 150 kHz) for tapping mode.
- Substrate: Sample firmly mounted on a 15 mm steel disc using double-sided adhesive.
Procedure:
- Mounting: Secure sample disc onto AFM piezoelectric scanner.
- Probe Engagement: Align laser, set photodetector sum to manufacturer's specification. Engage probe in contact or tapping mode using standard engage parameters.
- Scan Acquisition:
- Set scan size to a representative area (e.g., 20 µm x 20 µm to 100 µm x 100 µm).
- Set scan rate to 0.5-1.0 Hz for high resolution.
- Optimize feedback gains (Integral and Proportional) to minimize tracking error.
- Acquire scan in height sensor mode.
- Flattening & Analysis:
- Apply a 0th or 1st order flattening algorithm to the raw height image to remove tilt.
- Use the instrument's roughness analysis software.
- Define a primary profile from a single trace. Apply a Gaussian filter with a cutoff wavelength (λc) of 0.8 mm (for this profile length) to separate waviness from roughness.
- Calculate Ra (Arithmetic Mean Deviation) and Rq (Root Mean Square Deviation) from the roughness profile according to:
Ra = (1/L) ∫|Z(x)| dxRq = √[ (1/L) ∫ Z(x)² dx ]
- Report the mean and standard deviation from multiple (n≥3) scans on different sample regions.
Protocol 2: Nanomechanical Mapping via PeakForce QNM
Objective: To simultaneously map elastic modulus (E) and adhesion force with nanoscale resolution.
Materials:
- Sample: Hydrated or dry 3D-printed hydrogel (e.g., GelMA).
- AFM Probe: Silicon tip on a soft cantilever (e.g., Bruker ScanAsyst-Air, k ≈ 0.4 N/m). Pre-calibrate spring constant (thermal tune) and optical lever sensitivity.
- Fluid Cell: If measuring in liquid (PBS, media).
Procedure:
- Calibration: Perform a force curve on a clean, rigid reference sample (e.g., sapphire) to define the tip deflection sensitivity. Calibrate the tip radius using a characterized rough sample or after measurement via reconstruction.
- Engagement: Engage in PeakForce Tapping mode. Set PeakForce frequency to 0.25-2 kHz and amplitude to achieve desired contact force (~1-10 nN for soft materials).
- Mapping Acquisition: Set scan size and rate (typically 5-10 µm, 0.7 Hz). The system will acquire a force-distance curve at every pixel.
- Data Processing (Derrick Model Fit):
- For each curve, fit the retract portion to the Derjaguin-Muller-Toporov (DMT) model:
F = (4/3) E* √(R δ^(3/2)) + F_adh- Where
E*is the reduced modulus,Ris tip radius,δis indentation depth.
- The Adhesion Force is taken as the minimum force on the retract curve.
- The elastic modulus of the sample (
E_sample) is derived fromE*, assuming a known Poisson's ratio (νsample ~0.5 for hydrogels) and tip modulus (Etip):1/E* = (1-ν_sample²)/E_sample + (1-ν_tip²)/E_tip
- For each curve, fit the retract portion to the Derjaguin-Muller-Toporov (DMT) model:
- Reporting: Generate 2D maps of Modulus and Adhesion. Report mean values from homogenous regions, noting hydration state and loading rate.
Diagrams
Title: AFM Analysis Workflow for 3D Printed Materials
Title: Surface Parameters Influence Biological Response
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for AFM Surface Analysis of 3D-Printed Biomaterials
| Item | Function in Research | Example Product/Catalog |
|---|---|---|
| Standard AFM Calibration Grids | Verify lateral (µm-scale) and vertical (nm-scale) scanner accuracy. Essential for quantitative roughness. | Bruker PG: 1 µm pitch, 180 nm depth; BudgetSensors HS-100MG |
| Stiff Reference Sample | Calibrate force spectroscopy sensitivity and verify modulus measurement on a known standard. | Sapphire wafer, Fused Silica (E ~70 GPa) |
| Soft Reference Sample | Validate modulus measurements on compliant, hydrogel-like materials. | PDMS slabs of known modulus (e.g., 2 MPa), Polyacrylamide gels |
| Sharp AFM Probes (Si3N4) | For contact mode topography on soft polymers. Low spring constant minimizes sample damage. | Bruker DNP-S10, Olympus RC800PSA |
| PeakForce Tapping Probes | For high-res. nanomechanical mapping (PeakForce QNM). Pre-calibrated tips recommended. | Bruker ScanAsyst-Air (k~0.4 N/m), ScanAsyst-Fluid+ |
| Bio-Inert Liquid Cell | Enables AFM characterization in physiologically relevant buffers (PBS, cell culture media). | Bruker MTFML (for BioScope), JPK Liquid Pod |
| Sample Mounting Adhesive | Securely fix 3D-printed, often irregular, samples to AFM discs without contaminating surface. | Double-sided carbon tape, Blu-Tack reusable adhesive |
| Deionized Water & Solvents | For cleaning samples and probes. Isopropyl alcohol for degreasing, DI water for rinsing. | HPLC-grade isopropanol, 18.2 MΩ·cm DI water |
The Unique Advantages of AFM for Soft, Compliant, and Complex 3D Printed Structures
Atomic Force Microscopy (AFM) is an indispensable tool for the surface analysis of advanced 3D printed materials, particularly within the context of biomedical research and drug development. For soft, compliant, and geometrically complex 3D printed structures—such as tissue scaffolds, drug-eluting implants, and microfluidic devices—AFM provides unique advantages over other surface characterization techniques. Its ability to operate in fluid, apply minimal force, and map both topography and nanomechanical properties in three dimensions makes it uniquely suited for these challenging materials.
Recent studies, confirmed via current literature search, highlight AFM's critical role in quantifying the structure-function relationship of 3D printed biomaterials. Key application areas include:
- Nanomechanical Mapping: Quantifying the local elastic (Young's) modulus of hydrogel lattices and soft polymer scaffolds, correlating print parameters with mechanical compliance.
- Surface Morphology & Roughness: Precisely measuring the surface topography of printed features at the nano- and micro-scale, which influences cell adhesion and protein adsorption.
- Adhesion & Force Spectroscopy: Measuring binding forces between drug compounds and printed polymeric surfaces, or between cell receptors and functionalized scaffolds.
- Real-Time Monitoring: Observing structural degradation, swelling, or protein fouling on compliant printed surfaces in physiological buffers.
Key Experimental Protocols
Protocol 1: Nanomechanical Property Mapping of a 3D Printed Hydrogel Scaffold
Aim: To spatially map the Young's modulus of a poly(ethylene glycol) diacrylate (PEGDA) hydrogel lattice printed via digital light processing (DLP).
Materials:
- AFM with quantitative nanomechanical mapping (QNM) or PeakForce Tapping capability.
- Soft, compliant cantilever (nominal spring constant: 0.1 - 0.5 N/m, tip radius: < 20 nm).
- Calibration sample (e.g., polystyrene/polyethylene blend with known modulus).
- 3D printed PEGDA hydrogel sample in phosphate-buffered saline (PBS).
- Fluid cell or compatible liquid chamber.
Methodology:
- Cantilever Calibration: In air, determine the precise spring constant (k) using the thermal tune method. Calibrate the optical lever sensitivity (InvOLS) on a rigid surface in PBS.
- Tip Characterization: Image a characterized, sharp grating to determine the effective tip radius.
- Sample Preparation: Immobilize the hydrated hydrogel scaffold on a glass-bottom Petri dish using a thin layer of cyanoacrylate glue at the edges. Flood with PBS to prevent dehydration.
- AFM Mounting: Mount the dish on the scanner and engage the cantilever in liquid.
- Force Curve Acquisition: Set parameters for PeakForce Tapping mode: Peak Force Frequency = 0.25-1 kHz, Peak Force Setpoint = 100-500 pN (to limit strain to <10%).
- Mapping: Acquire a 50 µm x 50 µm map at 256 x 256 pixels resolution. The DMT or Sneddon model is applied to each force curve in real-time to calculate Young's modulus.
- Data Analysis: Use the instrument software to generate spatial modulus maps, histogram distributions, and correlate modulus with topographic features (e.g., strut junctions vs. center).
Protocol 2: Surface Roughness Analysis of a Drug-Loaded Polymeric Filament
Aim: To quantify the surface roughness (Sa, Sq) of a polycaprolactone (PCL) filament printed via fused deposition modeling (FDM), before and after drug (e.g., Doxycycline) incorporation.
Materials:
- AFM with tapping or contact mode capability.
- Silicon cantilever (nominal frequency: 150-300 kHz, spring constant: 5-40 N/m).
- Flat, rigid sample mounting discs.
- Double-sided adhesive tape.
- PCL and drug-loaded PCL printed specimens.
Methodology:
- Sample Mounting: Securely attach a small, flat section of the printed filament to a mounting disc using double-sided tape to minimize wobble.
- Cantilever Selection: Engage a stiff, sharp silicon tip for high-resolution topography.
- Imaging: In tapping mode, scan multiple 10 µm x 10 µm and 2 µm x 2 µm areas across the sample surface. Maintain a consistent scan rate (0.5-1 Hz) and setpoint ratio.
- Image Processing: Flatten scan lines to remove tilt. Apply a low-pass filter if necessary to remove high-frequency noise.
- Roughness Analysis: Use the software's roughness analysis tool to calculate the arithmetic mean height (Sa) and the root mean square height (Sq) for each scan. Compare values between pure and drug-loaded PCL surfaces.
Table 1: Comparison of Surface Characterization Techniques for Soft 3D Printed Structures
| Technique | Spatial Resolution (Lateral) | Mechanical Property Mapping | Measurement Environment | Sample Preparation Complexity | Key Limitation for Soft Materials |
|---|---|---|---|---|---|
| Atomic Force Microscopy (AFM) | ~0.5 nm | Yes (Quantitative) | Air, Liquid, Controlled Gas | Low | Scan size limited (<100µm typical) |
| Scanning Electron Microscopy (SEM) | ~1 nm | No | High Vacuum (typically) | High (coating, drying) | Not for hydrated samples; conductive coating alters surface |
| Optical Profilometry | ~0.2 µm | No (topography only) | Air | Low | Low lateral resolution; poor with transparent/compliant materials |
| Confocal Microscopy | ~0.2 µm | No (topography/fluorescence) | Air, Liquid | Medium | Limited to optical contrast; indirect mechanical data |
Table 2: Typical AFM-Derived Quantitative Data from 3D Printed Soft Materials
| Material | Printing Technique | AFM Mode | Key Measured Parameter | Representative Value Range | Biological/Functional Relevance |
|---|---|---|---|---|---|
| PEGDA Hydrogel | Digital Light Processing (DLP) | PeakForce QNM | Young's Modulus (E) | 5 - 50 kPa | Mimics soft tissue stiffness (e.g., brain, fat) |
| Alginate/Gelatin Bioink | Extrusion Bioprinting | Force Spectroscopy | Adhesion Force (Cell-Bioink) | 50 - 500 pN | Predicts cell attachment and spreading |
| Polycaprolactone (PCL) | Fused Deposition Modeling (FDM) | Tapping Mode | Surface Roughness (Sa) | 100 - 500 nm | Influences protein adsorption and osteointegration |
| PDMS Microfluidic Device | Stereolithography (SLA) | Contact Mode | Friction Coefficient | 0.1 - 0.5 | Determines fluid flow and cell shear stress |
Diagrams
AFM Analysis Workflow for 3D Printed Materials
AFM Data Informs Biomaterial Function
The Scientist's Toolkit: Key Research Reagent Solutions
| Item | Function in AFM Analysis of 3D Printed Structures |
|---|---|
| Functionalized AFM Tips (e.g., Collagen, RGD peptide) | Covalently modified tips measure specific biomolecular interaction forces between the printed surface and proteins or simulated cell membranes. |
| Calibration Gratings (TGF11, HS-100MG) | Essential for verifying the lateral accuracy of the AFM scanner and characterizing the geometry of the AFM tip itself. |
| Soft Cantilevers for QNM (e.g., ScanAsyst-Fluid+) | Silicon nitride tips on very flexible levers enable high-resolution, low-force imaging and modulus mapping of hydrogels in liquid. |
| Stiff Cantilevers for Tapping (e.g., RTESPA-300) | High-frequency, stiff silicon tips for high-resolution topographic imaging of stiffer polymers (e.g., PCL, PLA) with minimal surface damage. |
| Bio-Inert Liquid Cell | Allows stable imaging and force measurements in physiologically relevant buffers (PBS, cell culture medium) without contaminating the scanner. |
| Sample Mounting Adhesive (e.g., CrystalBond 509) | Thermally reversible adhesive to securely mount small, irregularly shaped 3D printed specimens to AFM discs without damaging the surface. |
Within the broader thesis on Atomic Force Microscopy (AFM) for 3D printing material surface analysis, this document outlines the critical application notes and protocols for addressing the lack of standardization in surface metrology for Additive Manufacturing (AM). The inherent layer-by-layer process of AM creates complex surfaces with unique topography, roughness, and texture that are not adequately captured by traditional 2D profilometry. The absence of standardized measurement protocols, parameters, and data analysis methods hinders reproducibility, quality control, and the correlation of surface features with functional performance, especially in regulated fields like biomedical device and drug delivery implant development.
The primary challenges in AM surface metrology stem from the multi-scale nature of AM surfaces, ranging from macro-scale warpage to nano-scale powder sinter features.
Table 1: Key Metrology Gaps and Their Impact on AM Research & Development
| Gap/Challenge | Description | Impact on Research/Development |
|---|---|---|
| Parameter Selection | Inconsistent use of roughness parameters (Sa, Sq, Sz) and spatial parameters (Sal, Str). Lack of guidelines for parameter relevance to AM surface types (e.g., as-built upskin vs. downskin). | Prevents direct comparison between studies, obscures process-property relationships. |
| Measurement Protocol | No consensus on sampling area, measurement location/orientation, filtering (S-F, L-F), and data stitching for large areas. | Introduces operator-dependent variability, reduces data reliability. |
| Instrument & Method Limitations | Confocal microscopy struggles with high aspect ratio valleys; SEM is primarily qualitative; stylus profilometry may damage soft polymers. | Incomplete surface characterization, missing critical topographic data. |
| Data Analysis & Reporting | Non-standardized formats for data storage and reporting. Proprietary software algorithms yield different results from the same dataset. | Hampers data sharing, meta-analysis, and the establishment of certified reference materials. |
| Correlation to Function | Difficulty linking specific surface metrics (e.g., hybrid parameters) to in-vivo performance (osseointegration, bacterial adhesion) or fluid flow. | Slows the iterative design of functional surfaces for drug-eluting implants or lab-on-a-chip devices. |
Detailed Experimental Protocols for AFM-Based Surface Characterization
Protocol 1: Multi-Scale AFM Topography Acquisition for AM Polymer Surfaces
- Objective: To obtain quantitative, three-dimensional topographical data from a selective laser sintered (SLS) polyamide (PA12) component at the micro- to nano-scale.
- Thesis Context: This protocol provides the foundational, reproducible methodology for the thesis's core AFM analysis.
- Materials: As-built PA12 specimen, anti-static air duster, AFM with scanning probe (e.g., silicon cantilever, tip radius <10 nm).
- Procedure:
- Sample Preparation: Use compressed, oil-free air to remove loose powder particles. Mount the sample firmly on a magnetic or adhesive AFM disc. Avoid chemical cleaning unless its effect is part of the study.
- Probe Selection: Choose a non-contact or tapping mode probe appropriate for polymers (medium spring constant ~40 N/m) to minimize sample damage.
- Site Selection: Using an integrated optical microscope, identify and target three representative areas: an upskin (top) surface, a downskin (bottom) surface, and a vertical side surface. Mark locations.
- Scan Acquisition:
- Perform an initial large-area scan (e.g., 100 µm x 100 µm) to assess general topography.
- Subsequently, perform higher-resolution scans (e.g., 10 µm x 10 µm and 2 µm x 2 µm) at each location to capture finer details of powder particles and sinter necks.
- Set scan rate to 0.5-1 Hz to optimize signal-to-noise ratio.
- Apply real-time plane leveling (flattening order 1 or 2).
- Data Saving: Save raw data (.spm, .ibw, etc.) and export height, amplitude, and phase images in a standardized format (e.g., .tiff).
Protocol 2: Post-Processing and Roughness Parameter Calculation
- Objective: To standardize the derivation of ISO 25178 areal surface parameters from AFM data.
- Procedure:
- Import: Load the raw height data into a surface metrology software package (e.g., Gwyddion, MountainsMap).
- Form Removal: Apply a least-squares mean plane subtraction (flattening) to remove sample tilt.
- Filtering:
- Apply an S-Filter (Gaussian filter, λs = 0.25 µm) to remove high-frequency noise.
- Apply an L-Filter (Gaussian regression filter, λc = 8 µm) to separate roughness from waviness. The selection of λc must be reported.
- Masking: Manually or automatically mask out non-measurement artifacts (e.g., deep pits from entrapped powder).
- Parameter Extraction: Calculate and record the following core set of parameters for each scan:
- Height Parameters: Sa (Arithmetical mean height), Sq (Root mean square height), Sz (Maximum height).
- Spatial Parameters: Sal (Autocorrelation length), Str (Texture aspect ratio).
- Hybrid Parameters: Sdr (Developed interfacial area ratio).
Visualization of the Standardization Workflow and Challenge Relationships
Title: AM Surface Metrology Challenges & AFM Solution Path
Title: AFM Data Post-Processing Protocol Workflow
The Scientist's Toolkit: Research Reagent Solutions & Essential Materials
Table 2: Essential Toolkit for Standardized AFM Surface Metrology in AM Research
| Item | Function/Description | Application Note |
|---|---|---|
| AFM with Environmental Control | Enables scanning in non-contact/tapping mode to prevent damage to soft AM polymers. Humidity/temp control ensures measurement stability. | Critical for soft materials (e.g., PEEK, hydrogels). Use anti-vibration table. |
| Standardized Reference Sample | A physical artifact with known, traceable roughness values (e.g., ISO 5436-1). | Used for daily verification of AFM lateral and vertical calibration. |
| Surface Metrology Software (e.g., MountainsMap, Gwyddion) | Software compliant with ISO 25178 for areal surface parameter calculation with controlled filtering. | Avoid using instrument-default software only; ensures algorithmic consistency. |
| Stable Sample Mounting Kit | Includes magnetic disks, conductive tape, and adjustable sample stages. | Prevents sample drift during long or high-resolution scans. |
| Probe Kit for Diverse Materials | Includes high-resolution silicon probes for polymers, diamond-coated probes for metals/ceramics, and conductive probes. | Matching probe to material prevents damage and artifacts. |
| Data Format Standard (e.g., OPFS) | Open吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕吕 |
A Step-by-Step Guide to AFM Protocol for 3D Printed Polymer and Biomaterial Surfaces
Atomic Force Microscopy (AFM) analysis of 3D printed materials provides critical insights into surface topography, roughness, and mechanical properties, which are essential for evaluating print fidelity, layer adhesion, and post-processing effects. Within the broader thesis on AFM for 3D printing material surface analysis, proper specimen preparation is paramount. Inconsistent mounting, contamination, and improper handling are primary sources of artifacts that can compromise data integrity. These application notes detail standardized protocols to ensure reproducible, high-fidelity AFM measurements on polymeric, composite, and resin-based 3D printed specimens.
Mounting Protocols for Stability
Secure and stable mounting is critical to prevent vibration and drift during AFM scanning.
Protocol: Rigid Substrate Adhesion for Planar Specimens
- Objective: To immobilize a specimen with a flat base onto a standard AFM specimen disc.
- Materials: Double-sided adhesive tape (carbon-conductive or high-tack acrylic), cyanoacrylate gel, or two-part epoxy; flat AFM metal discs; forceps.
- Method:
- Clean the AFM disc with isopropanol and lint-free wipes. Allow to dry.
- For tape: Apply a piece slightly smaller than the specimen to the disc. Remove the liner and gently place the specimen onto the tape. Apply firm, even pressure for 30 seconds.
- For adhesive/gel: Apply a minimal dot to the center of the disc. Position the specimen and apply gentle pressure. Allow full curing per manufacturer instructions (e.g., 5 min for gel, 24 hrs for epoxy).
- Key Consideration: Adhesive choice depends on scan forces and required conductivity. Excess adhesive must be avoided to prevent contaminating the scan area.
Protocol: Custom Fixturing for Non-Planar or Fragile Specimens
- Objective: To mount irregularly shaped or delicate prints without inducing stress or instability.
- Materials: Custom 3D printed or machined clamps/holders, modeling clay (non-outgassing), vacuum chuck.
- Method:
- Design a fixture that supports the specimen at its base or sides, leaving the scan area completely accessible and unobstructed.
- Secure the fixture firmly to the AFM disc using an adhesive.
- Gently place the specimen into the fixture. If using compliant clay, embed only the very edges of the specimen's base.
- For porous specimens, a low-pressure vacuum chuck can be effective.
Cleaning Protocols for Surface Contaminant Removal
Residual support material, oils, and dust are common contaminants that obscure true surface morphology.
Protocol: Dry Cleaning for Particulate Removal
- Objective: To remove loose powder, dust, and support fragments without altering the surface.
- Materials: Clean, dry nitrogen or compressed air gun (with in-line 0.2 µm filter), soft anti-static brushes.
- Method:
- Hold the specimen at an angle. Using short, controlled bursts from a distance of 2-5 cm, direct the gas stream across the surface.
- For delicate surfaces, use a soft anti-static brush with gentle, sweeping motions.
- Always perform this step before any wet cleaning.
Protocol: Solvent Cleaning for Organic Residue Removal
- Objective: To dissolve and remove printing oils, uncured resin, or fingerprint residues.
- Critical Pre-Test: Solvent compatibility must be tested on a non-critical area of the print material to check for swelling, cracking, or dissolution.
- Method:
- Immerse or gently flood the specimen surface with the appropriate solvent.
- Agitate gently via low-power ultrasonication (<40 kHz) for 30-60 seconds if the specimen is not fragile.
- Rinse immediately with a second aliquot of clean solvent.
- Dry thoroughly under a filtered nitrogen stream.
Table 1: Recommended Cleaning Solvents for Common 3D Print Materials
| Material Class (Example) | Recommended Solvent(s) | Contraindicated Solvents | Application Notes |
|---|---|---|---|
| Photopolymers (SLA, DLP Resins) | Isopropyl Alcohol (IPA), Ethanol | Acetone, Chlorinated solvents | IPA immersion followed by nitrogen dry is standard for uncured resin removal. |
| Fused Filaments (ABS) | Acetone (for vapor smoothing), IPA | N/A | Acetone will aggressively smooth; use IPA for gentle cleaning. |
| Fused Filaments (PLA) | IPA, Ethanol | Acetone | Acetone can degrade PLA surface. |
| Polyjet (Stratasys) Photopolymers | Water, diluted detergent, IPA | Strong organic solvents | Support material is water-soluble; follow manufacturer guidelines. |
| SLS Nylon (PA11, PA12) | Isopropyl Alcohol | Acetone (can cause stress cracking) | Effective for removing loose powder. |
Handling and Storage Protocols
Proper handling minimizes introduction of new contaminants or damage prior to analysis.
Protocol: Contamination-Minimized Handling
- Objective: To transfer and position specimens without depositing particulates or oils.
- Materials: Powder-free nitrile gloves, clean forceps with non-scratching tips (e.g., plastic or coated), dedicated clean storage containers.
- Method:
- Always wear clean gloves. Handle specimens by their edges or non-analysis areas only.
- Use forceps for final positioning on the AFM stage.
- Never touch the analysis surface or the AFM probe.
Protocol: Short- & Long-Term Storage
- Objective: To preserve specimen surface state between printing and measurement.
- Materials: Clean, sealed glass or plastic containers, desiccant packets.
- Short-Term (<24 hrs): Store in a covered, clean Petri dish or similar.
- Long-Term (>24 hrs): Place in an airtight container with desiccant. Store in dark, temperature-stable conditions to prevent material creep or UV degradation.
Workflow & Experimental Pathway
AFM Sample Preparation Workflow for 3D Prints
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 2: Key Reagents and Materials for AFM Sample Preparation
| Item | Function in Protocol | Key Consideration |
|---|---|---|
| High-Purity Isopropanol (IPA) | Primary solvent for cleaning photopolymer resins and many filaments. Removes organic residues. | Use HPLC or electronic grade to avoid non-volatile impurities. |
| Filtered, Dry Nitrogen Gun | Dust-free drying after cleaning; dry particulate removal. | In-line 0.2 µm filter is mandatory to prevent oil/particulate deposition. |
| Double-Sided Carbon Tape | Conductive mounting for planar specimens. Prevents charging in electrical modes. | Ensures electrical contact for conductive-AFM or Kelvin Probe modes. |
| Cyanoacrylate Gel Adhesive | Rigid, fast-curing mounting for unstable or tall specimens. | Gel formulation minimizes wicking to the analysis surface. |
| Powder-Free Nitrile Gloves | Mandatory for handling to prevent skin oil and salt contamination. | Latex gloves can deposit particulates; cotton gloves can shed fibers. |
| Anti-Static Brushes | Gentle removal of electrostatic dust from fragile surfaces. | Natural soft hair (e.g., camel) is preferred to prevent scratching. |
| Clean Room Wipes (Lint-Free) | Wiping AFM discs and tools with solvent. | Non-woven polyester or cellulose are suitable low-lint options. |
| Desiccant (Silica Gel) | Maintaining dry storage atmosphere to prevent hydrolysis or creep. | Use indicator beads and regenerate regularly. |
Within the broader thesis on Atomic Force Microscopy (AFM) for 3D printing material surface analysis, selecting the appropriate imaging mode is critical for obtaining accurate, high-resolution topographical and mechanical property data. This application note provides detailed protocols and comparisons for Contact, Tapping, and PeakForce Tapping modes, tailored for researchers analyzing advanced polymers, composites, and bio-printed materials used in scientific and drug development applications.
Key AFM Mode Characteristics & Quantitative Comparison
The following table summarizes the core operational parameters and optimal application ranges for each mode, based on current instrument specifications and published research.
Table 1: Quantitative Comparison of Primary AFM Modes for Material Analysis
| Parameter | Contact Mode | Tapping Mode | PeakForce Tapping Mode |
|---|---|---|---|
| Tip-Sample Interaction | Constant physical contact | Intermittent contact (oscillating) | Pulsed, sub-100 pN to >10 nN force |
| Typical Force Applied | 0.5 - 100 nN | 0.1 - 5 nN (peak force) | Precisely controlled, often <1 nN |
| Lateral (Shear) Forces | High | Negligible | Very Low |
| Imaging Speed | Moderate | Fast | Fast (with quantitative data) |
| Best Vertical Resolution | <0.1 nm | ~0.1 nm | ~0.1 nm |
| Sample Damage Risk | High (soft samples) | Moderate to Low | Very Low |
| Key Measured Properties | Topography, Friction | Topography, Phase (adhesion/viscoelasticity) | Topography, Young's Modulus, Adhesion, Deformation, Dissipation |
| Ideal Material Types | Hard, flat, inert surfaces (e.g., silicon, metals) | Soft polymers, biological samples, heterogeneous surfaces | All, especially: ultra-soft gels, compliant polymers, multicomponent 3D prints |
Detailed Experimental Protocols
Protocol 1: Contact Mode Imaging for Hard 3D-Printed Resins
Objective: To obtain high-resolution topography of rigid, cured photopolymer resin surfaces. Materials: AFM with contact mode scanner, Si or Si3N4 contact mode probes (k ~ 0.2 N/m), rigid 3D-printed sample. Procedure:
- Probe & Laser Alignment: Mount a contact mode cantilever. Align the laser spot on the cantilever's end and adjust the photodetector to achieve a sum signal maximum and a vertical deflection (VD) signal near zero.
- Engagement: Position the tip above a featureless area of the sample. Initiate automatic engagement with a setpoint force of ~5-10 nN.
- Feedback Optimization: Set scan parameters (Scan rate: 1-2 Hz, Scan size: 5x5 μm). Adjust the Proportional (P) and Integral (I) gains to maintain a setpoint with minimal oscillation in the VD signal.
- Data Acquisition: Acquire topography and lateral force (friction) images simultaneously. Save raw data channels.
- Post-Processing: Apply a first-order flattening to remove sample tilt. Analyze surface roughness (Ra, Rq).
Protocol 2: Tapping Mode Imaging for Polymer Blend Phase Separation
Objective: To map topography and phase distribution in a thermoplastic polyurethane/polycarbonate blend. Materials: AFM with tapping mode capability, stiff tapping probe (k ~ 40 N/m, f0 ~ 300 kHz), polymer blend sample. Procedure:
- Tune Cantilever: In air, auto-tune the cantilever to find its resonant frequency (f0) and quality factor (Q).
- Set Parameters: Set drive amplitude (A_drive) to 50-80% of the free amplitude. Choose a scan rate of 0.5-1 Hz.
- Engage & Setpoint: Engage with a setpoint amplitude (A_sp) of ~70-80% of the free amplitude.
- Optimize Feedback: Adjust P and I gains to maintain A_sp without ringing. The Phase signal should be monitored simultaneously.
- Acquire Data: Collect Topography and Phase images. The phase contrast reveals domains with differing adhesive or viscoelastic properties.
- Analysis: Use histogram analysis of the phase image to quantify area fraction of distinct domains.
Protocol 3: PeakForce Tapping Nanomechanical Mapping of a Bio-Ink
Objective: To quantitatively map the modulus and adhesion of an alginate-based bio-printed hydrogel. Materials: AFM with PeakForce QNM capability, sharp silicon probe with calibrated spring constant (k ~ 0.7 N/m) and tip radius, bio-ink sample in hydrated cell culture medium. Procedure:
- Probe Calibration: Perform thermal tune to determine k and deflection sensitivity. Input known tip radius for modulus quantification.
- Fluid Setup: Mount sample in fluid cell. Submerge tip. Re-align laser and re-tune in fluid (lower f0, higher Q).
- Enable PeakForce Tapping: Set PeakForce frequency to 1-2 kHz. Set a low PeakForce amplitude (~50-100 nm).
- Optimize PeakForce Setpoint: Adjust the peak force setpoint to the lowest value that maintains stable imaging (~100-500 pN for gels).
- Acquire Quantitative Maps: Collect Topography, Young's Modulus (Derjaguin–Müller–Toporov (DMT) model), Adhesion, and Deformation maps simultaneously.
- Data Validation: Ensure modulus values are within the model's applicability for the sample. Use a reference sample of known modulus for verification.
AFM Mode Selection Workflow Diagram
Title: AFM Mode Selection Decision Tree
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 2: Key Materials for AFM Analysis of 3D-Printed Surfaces
| Item | Function & Relevance |
|---|---|
| Si3N4 Contact Probes (k ~ 0.1 N/m) | Low spring constant for imaging hard materials with minimal induced deformation in Contact Mode. |
| High-Frequency Si Tapping Probes (k ~ 40 N/m, f0 ~ 300 kHz) | Stiff probes for high-resolution Tapping Mode in air; phase imaging for material contrast. |
| SCANASYST-FLUID+ Probes | Proprietary probes optimized for PeakForce Tapping in fluid; pre-calibrated for consistent nanomechanical data. |
| Polystyrene-Polyethylene (PS-PE) Reference Sample | Standard sample with known, distinct domains for verifying Tapping Mode phase contrast and tip condition. |
| PDMS Calibration Grid | Sample with periodic structures of known height and pitch for verifying AFM scanner calibration in X, Y, and Z. |
| Deionized Water & HPLC-Grade Ethanol | For cleaning substrates and probes, and for imaging in liquid environments to reduce capillary forces. |
| Adhesive Tape or Thermal Conductive Paste | For securely mounting small or irregular 3D-printed samples to AFM specimen disks to prevent drift. |
| Argon Gas Duster | For removing particulate contamination from samples and the AFM stage without contact. |
Within the context of Atomic Force Microscopy (AFM) applied to 3D printing material surface analysis, precise optimization of operational parameters is critical for generating reliable, high-fidelity nanoscale topography data. This data directly informs research on surface roughness, layer adhesion, and drug release kinetics from printed pharmaceutical formulations. This document provides application notes and protocols for methodically optimizing three interdependent parameters: Scan Rate, Resolution (pixels per line), and Setpoint, to balance imaging quality, tip integrity, and data acquisition efficiency.
The Parameter Triad: Core Concepts & Interdependencies
Scan Rate
The speed at which the probe tip travels across the sample surface, typically measured in Hz (lines per second). Excessively high rates can cause tip skipping or deformation of soft materials, while low rates increase scan time and drift susceptibility.
Resolution (Pixel Density)
Defined by the number of data points sampled per line (X-resolution) and the number of lines per image (Y-resolution). Higher resolution reveals finer detail but requires slower scan rates or risks oversampling.
Setpoint
The target value for the feedback loop (e.g., oscillation amplitude in tapping mode, deflection in contact mode). It defines the tip-sample interaction force. A low setpoint increases force, potentially damaging soft samples; a high setpoint risks instability and loss of contact.
Interdependency
These parameters are intrinsically linked. Increasing resolution necessitates a proportional decrease in scan rate to maintain data point sampling time. The setpoint must be adjusted relative to the scan rate to ensure the feedback loop can track topography accurately at the chosen speed.
Quantitative Parameter Guidelines for 3D Printing Materials
The following table summarizes recommended starting parameters and optimization ranges for common 3D printed material classes, derived from current literature and experimental practice.
Table 1: Initial AFM Parameter Ranges for 3D Printed Material Classes
| Material Class (Example) | Recommended Mode | Initial Setpoint Ratio* | Initial Scan Rate (Hz) | Recommended Resolution (pixels) | Key Consideration |
|---|---|---|---|---|---|
| Hard Thermoplastics (PLA, ABS) | Tapping Mode | 0.7 - 0.8 | 1.0 - 2.0 | 512 x 512 | High setpoint for durability; moderate rate for layer edge definition. |
| Flexible Polymers/Elastomers (TPU, Silicones) | Tapping Mode (Low Amp) | 0.85 - 0.95 | 0.5 - 1.0 | 512 x 512 or 256 x 256 | High setpoint ratio minimizes force; slow scan to prevent surface deformation. |
| Photopolymer Resins (SLA/DLP Printed) | Tapping Mode | 0.75 - 0.85 | 0.8 - 1.5 | 512 x 512 | Potential for tip contamination; medium setpoint balances tracking and safety. |
| Pharmaceutical Blends (API-Polymer Matrices) | Tapping Mode | 0.80 - 0.90 | 0.3 - 0.7 | 1024 x 1024 | Very soft; ultra-low force and slow scan essential to resolve API crystals. |
| Hydrogels/Bioprinted | Tapping Mode in Fluid | 0.90 - 0.98 | 0.1 - 0.5 | 256 x 256 | Near-free amplitude operation; extremely slow scanning in liquid. |
| Metallic/Composite | Contact Mode | 0.5 - 2.0 nA | 1.0 - 3.0 | 512 x 512 | Stable deflection setpoint; higher rates possible on hard surfaces. |
Ratio of setpoint amplitude to free-air oscillation amplitude. *Deflection setpoint in nA or V.
Experimental Protocol: Iterative Parameter Optimization
Protocol 1: Establishing Baseline Conditions for a Novel Material
Objective: To determine a stable, non-destructive starting point for imaging an unknown 3D printed sample. Materials: AFM with tapping mode capability; appropriate cantilever; 3D printed sample; calibration grating. Procedure:
- Cantilever Tuning: Resonate the cantilever in air (or fluid). Note the free amplitude (A~0~).
- Coarse Approach: Engage on a representative, feature-rich area of the sample using conservative auto-approach settings.
- Initial Safe Parameters: Set scan size to 5 µm, resolution to 256 x 256, scan rate to 0.5 Hz.
- High Setpoint Engage: Set the amplitude setpoint to 90% of A~0~. Engage the feedback loop.
- Initial Scan & Observation: Start scanning. Observe the trace/retrace error signal and the topography image in real-time.
- Lower Setpoint Iteratively: Gradually decrease the setpoint (e.g., to 80%, then 70%) while monitoring:
- Image Quality: Does topographic detail improve?
- Error Signal: Does it become excessively noisy or smooth out?
- Probe Integrity: Listen for audible tapping changes indicating contamination or damage.
- Define Optimal Setpoint: Identify the lowest setpoint where the error signal remains stable and trace/retrace overlay is good. This is the Baseline Setpoint (S~b~).
Protocol 2: Optimizing Scan Rate & Resolution at Baseline Setpoint
Objective: To maximize image quality and acquisition speed without losing tracking or damaging the sample. Materials: AFM system with sample engaged at S~b~. Procedure:
- Fix Parameters: Lock the setpoint at S~b~. Select a 1-2 µm scan area with distinct features.
- Test Resolution Suite: Acquire images at a fixed, slow scan rate (0.3 Hz) with increasing resolution:
- Sequence: 128 x 128 → 256 x 256 → 512 x 512 → 1024 x 1024.
- Assess: Determine the resolution where finer details (e.g., polymer spherulites, nano-pores) cease to improve. This is the Useful Resolution (R~u~).
- Optimize Scan Rate: Using R~u~, systematically increase the scan rate:
- Sequence: 0.3 Hz → 0.5 Hz → 0.8 Hz → 1.2 Hz → 2.0 Hz.
- Evaluate at Each Rate: Capture images and analyze:
- Topographic Integrity: Compare to the 0.3 Hz baseline. Look for smearing, elongation, or loss of sharp edges.
- Error Signal: Check for increasing noise or feedback loop oscillations.
- Line Profile Consistency: Measure the height/width of a consistent feature across rates.
- Define Maximum Reliable Rate (R~max~): The highest scan rate before a >10% distortion in feature dimensions or a significant loss of signal-to-noise occurs.
Table 2: Optimization Decision Matrix (Post-Protocol Analysis)
| Observed Artifact | Probable Cause | Corrective Action |
|---|---|---|
| Smearing/Elongation | Scan rate too high for feedback response. | Decrease scan rate by 30-50%. |
| Excessive Noise/Grain | Setpoint too low or rate too high. | Slightly increase setpoint ratio or decrease scan rate. |
| Flat/Featureless Image | Setpoint too high (skimming). | Decrease setpoint ratio incrementally. |
| Horizontal Striping | Low scan rate with high resolution causing drift. | Increase scan rate slightly or decrease resolution. |
| Asymmetric Features | Contaminated tip or scanner hysteresis. | Change tip, perform scanner calibration. |
Visualization of the Optimization Workflow
Diagram 1: AFM Parameter Optimization Workflow
Diagram 2: Core Parameter Interdependencies
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 3: Essential AFM Toolkit for 3D Printing Material Analysis
| Item | Function/Explanation | Critical for Material Class |
|---|---|---|
| PPP-FMR Cantilevers | Tapping mode probes with reflex coating and very sharp tip (~2 nm radius). Essential for high-resolution imaging of polymer surfaces and nano-features. | All, especially hard thermoplastics & resins. |
| PNP-DB Cantilevers | Self-actuating, self-sensing probes for conductive samples. Used for electrical property mapping of composite or doped printed materials. | Conductive composites, printed electronics. |
| SCANASYST-FLUID+ Probes | Optimized for tapping mode in liquid with very soft springs. Critical for hydrated hydrogel and bioprinted structure analysis. | Hydrogels, bioprinted scaffolds. |
| OTR8 Thermal Oxide Calibration Grating | Provides absolute Z-height and XY spatial calibration (8 µm pitch). Verifies scanner linearity and measurement accuracy post-optimization. | All (regular calibration). |
| Adhesive Tape/Clean Wafers | For secure, flat mounting of irregular 3D printed samples to magnetic AFM disks. Minimizes sample wobble. | All, especially rough prints. |
| UV-Ozone Cleaner | Removes organic contaminants from silicon tips and sample surfaces, reducing artifacts and improving reproducibility. | All, prior to critical experiments. |
| Deionized Water & IPA | For cleaning samples (removing loose powder, residue) and cantilever holders. Essential for preventing tip contamination. | All. |
| Nitrogen Gas Duster | For dry, non-contact cleaning of samples and the AFM stage to remove environmental dust particles. | All. |
Quantifying Surface Roughness and Pore Structure of 3D Printed Scaffolds
Within the broader thesis on Atomic Force Microscopy (AFM) for 3D printing material surface analysis, this Application Note details the quantitative characterization of 3D printed scaffolds. For tissue engineering and drug delivery applications, surface topography (roughness) and pore architecture (size, distribution, interconnectivity) are critical parameters influencing cell adhesion, proliferation, differentiation, and drug release kinetics. This document provides standardized protocols for AFM-based surface roughness quantification and complementary image analysis for pore structure assessment.
Key Research Reagent Solutions
The following table lists essential materials and reagents commonly employed in the preparation and analysis of 3D printed scaffolds for such studies.
| Item Name | Function/Brief Explanation |
|---|---|
| Polylactic Acid (PLA) | A biodegradable thermoplastic polymer, widely used for fabricating bone and tissue scaffolds via Fused Deposition Modeling (FDM). |
| Polycaprolactone (PCL) | A semi-crystalline, bioresorbable polyester with a low melting point, suitable for extrusion-based 3D printing and long-term implant studies. |
| Gelatin Methacryloyl (GelMA) | A photo-crosslinkable hydrogel bioink for bioprinting; mimics the extracellular matrix, allowing cell encapsulation and pore formation. |
| Phosphate Buffered Saline (PBS) | Used for hydrating and rinsing hydrogel scaffolds to maintain physiological pH and ion concentration during imaging or testing. |
| Critical Point Dryer | Equipment used to dry hydrated or soft polymeric scaffolds without collapsing delicate pore structures by avoiding liquid-vapor interfaces. |
| Conductive Tape/Sputter Coater | For non-conductive scaffolds, a thin metal coating (e.g., gold) is applied prior to SEM imaging to prevent charging and improve image quality. |
| Nanoindentation AFM Probe (e.g., RTESPA-300) | A stiff cantilever with a sharp tip for high-resolution topography mapping and nanomechanical property measurement of scaffold surfaces. |
Experimental Protocols
Protocol 1: AFM-Based Surface Roughness Quantification
Objective: To obtain high-resolution, quantitative topographical data from the strut surfaces of 3D printed scaffolds.
Materials & Equipment:
- Atomic Force Microscope (e.g., Bruker Dimension Icon, Cypher ES)
- Soft Contact Mode or Tapping Mode AFM probe (spring constant ~0.4-4 N/m, resonant frequency ~60-90 kHz in air)
- 3D printed scaffold sample (dry or hydrated as required)
- Sample mounting discs and adhesive
- Software: Nanoscope Analysis, Gwyddion, or SPIP.
Methodology:
- Sample Preparation: Securely mount a small section of the scaffold (~5x5 mm) onto a metal specimen disc using a small amount of adhesive (e.g., double-sided tape or cyanoacrylate). Ensure the surface of interest is level and accessible. For hydrogels, perform measurements in fluid cells with appropriate buffer.
- Probe Selection & Mounting: Choose a sharp silicon nitride or silicon tip appropriate for the sample's stiffness. Mount the probe and perform laser alignment and photodetector adjustment.
- Engagement: Position the probe above a representative, flat region of a scaffold strut. Engage the tip onto the surface using standard engagement procedures.
- Scanning Parameters: Set a scan size typically between 10x10 µm and 50x50 µm to capture relevant micro-features. Use a slow scan rate (e.g., 0.5-1.0 Hz) for optimal resolution. Maintain a setpoint to ensure minimal, non-destructive force.
- Data Acquisition: Acquire height, amplitude, and phase images. Capture at least 5 images from different struts across the scaffold to ensure statistical significance.
- Image Processing & Analysis:
- Flatten or plane-fit each height image to remove background tilt.
- Apply a noise filter if necessary.
- Use the software's roughness analysis toolbox to calculate the following parameters:
- Ra (Average Roughness): The arithmetic average of absolute deviations from the mean plane.
- Rq (Root Mean Square Roughness): The standard deviation of height values.
- Rz (Average Maximum Height): The average difference between the five highest peaks and five lowest valleys.
- Surface Area Ratio: The ratio of the 3D surface area to the 2D projected area.
Protocol 2: Pore Structure Analysis via SEM/µCT Image Processing
Objective: To quantify pore geometry and network architecture from cross-sectional images.
Materials & Equipment:
- Scanning Electron Microscope (SEM) or Micro-Computed Tomography (µCT) scanner
- ImageJ/FIJI software with BoneJ plugin or equivalent (e.g., CTAn for µCT)
Methodology (for SEM-derived 2D analysis):
- Image Acquisition: Capture high-contrast, top-down and cross-sectional SEM images of the scaffold at consistent magnification (e.g., 100x, 500x). Ensure scale bars are included.
- Image Pre-processing (in ImageJ):
- Convert image to 8-bit grayscale.
- Adjust threshold (using Otsu's method or manual setting) to create a binary image where pores are black and material is white.
- Apply "Fill Holes" and "Remove Outliers" functions to clean the binary mask.
- Quantitative Analysis (using BoneJ plugin or built-in functions):
- Pore Size (Diameter): Use "Analyze Particles" function. Set a circularity limit (e.g., 0.4-1.0) to exclude non-pore artifacts. Record pore area and calculate equivalent circular diameter.
- Porosity: Calculate as the percentage of black pixels (pores) relative to total pixels in the region of interest (ROI).
- Pore Interconnectivity (2D Approximation): Skeletonize the binary image (Process > Binary > Skeletonize). Analyze the skeleton to count branch points and assess network connectivity.
Data Presentation
Table 1: Representative AFM Surface Roughness Parameters of Various 3D Printed Scaffold Materials
| Material | Printing Technique | Ra (nm) | Rq (nm) | Rz (nm) | Surface Area Ratio | Reference Condition |
|---|---|---|---|---|---|---|
| PCL | Melt Electrowriting (MEW) | 45 ± 12 | 58 ± 15 | 320 ± 45 | 1.08 ± 0.02 | Dry, as-printed |
| PLA | Fused Deposition Modeling (FDM) | 520 ± 85 | 660 ± 110 | 3100 ± 600 | 1.32 ± 0.05 | Dry, as-printed |
| GelMA | Digital Light Processing (DLP) | 18 ± 5 | 24 ± 7 | 150 ± 30 | 1.01 ± 0.01 | Hydrated (PBS) |
| PLA with Surface Etching (NaOH) | FDM + Post-process | 1200 ± 200 | 1450 ± 250 | 8500 ± 1200 | 1.85 ± 0.12 | Dry |
Table 2: Pore Structure Metrics from Image Analysis of Scaffolds
| Material | Mean Pore Size (µm) | Porosity (%) | Pore Circularity (0-1) | Interconnectivity (Branch Points/mm²) | Analysis Method |
|---|---|---|---|---|---|
| PCL (MEW) | 25 ± 5 | 65 ± 4 | 0.85 ± 0.08 | 1200 ± 150 | SEM 2D |
| β-TCP Ceramic | 350 ± 50 | 75 ± 3 | 0.65 ± 0.10 | 250 ± 40 | µCT 3D |
| Collagen-GAG | 160 ± 20 | 98 ± 1 | 0.55 ± 0.15 | Highly interconnected | SEM 2D |
| PLA (FDM, 0/90° laydown) | 400 x 400 (square) | 45 ± 2 | 0.95 ± 0.05 | 40 ± 10 | Optical Microscopy |
Visualization
Workflow for Scaffold Roughness and Pore Analysis
How Scaffold Topography Influences Cell & Drug Response
This application note, framed within the broader thesis of utilizing Atomic Force Microscopy (AFM) for the surface analysis of 3D-printed biomedical materials, details the quantitative mapping of nanomechanical properties critical for material performance. For 3D-printed hydrogels and composites used in drug delivery, tissue engineering, and biosensing, elasticity (Young's modulus) and adhesion forces are not bulk averages but spatially heterogeneous properties that dictate cell-material interactions, drug release kinetics, and structural integrity. AFM-based nanomechanical mapping is indispensable for correlating print parameters (e.g., layer height, curing intensity, bioink composition) with local functional properties at the micro- and nanoscale, providing feedback unattainable by bulk rheology.
Table 1: Representative Nanomechanical Properties of 3D-Printed Hydrogels & Composites
| Material System | Printing Method | Average Young's Modulus (kPa) | Adhesion Force (nN) | Spatial Resolution (nm) | Key Application Context |
|---|---|---|---|---|---|
| GelMA Hydrogel | Digital Light Processing (DLP) | 12.5 ± 3.2 | 0.25 ± 0.08 | 50 | Soft tissue scaffolds, cell mechanobiology studies |
| Alginate-Polyacrylamide Dual Network | Extrusion-based | 85.4 ± 12.7 | 1.8 ± 0.4 | 100 | Load-bearing osteochondral constructs |
| PEGDA-Silica Nanoparticle Composite | Stereolithography (SLA) | 1,250 ± 210 | 5.5 ± 1.2 | 80 | Stiff, abrasion-resistant dental guides |
| Collagen-Hyaluronic Acid Bioink | Extrusion (Bioprinting) | 8.1 ± 2.1 | 2.3 ± 0.6 | 150 | Skin regeneration models, drug penetration assays |
| PLA-PEG Blend (Surface) | Fused Deposition Modeling (FDM) | 2.1 x 10^6 ± 0.3 x 10^6 | 15.0 ± 3.5 | 20 | Drug-eluting implant coating durability |
Data synthesized from recent literature (2023-2024) on AFM analysis of printed soft materials.
Experimental Protocols
Protocol 1: AFM Nanomechanical Mapping via PeakForce QNM Objective: To simultaneously map elastic modulus and adhesion of a 3D-printed hydrogel surface in a physiologically relevant fluid. Materials: See "The Scientist's Toolkit" below. Procedure:
- Sample Preparation: Hydrate the 3D-printed sample in PBS (pH 7.4) for 1 hour prior to mounting. Secure sample to a magnetic AFM dish using a thin layer of cyanoacrylate adhesive at edges only, ensuring the surface is level.
- Cantilever Selection & Calibration: Use a silicon nitride cantilever with a nominal spring constant (k) of 0.1-0.6 N/m and a spherical silica tip (diameter 2-5 µm). Perform thermal tuning in fluid to determine the exact k value. Determine the optical lever sensitivity (InvOLS) on a clean, rigid sapphire surface in PBS.
- Parameter Optimization: Set the PeakForce frequency to 0.5-1 kHz and amplitude to 100-150 nm. Adjust the PeakForce Setpoint to achieve a consistent contact force (~1 nN for soft gels). Enable quantitative nanomechanical property mapping mode.
- Data Acquisition: Perform a 50 µm x 50 µm scan at 256 x 256 pixels resolution. Collect data for Height, Young's Modulus (DMT model), and Adhesion force channels simultaneously.
- Data Processing: Apply a first-order plane fit to height data. Use dedicated software (e.g., NanoScope Analysis) to generate modulus and adhesion maps. Apply necessary filters (e.g., flatten) to property maps, excluding pixels from obvious debris.
Protocol 2: Adhesion Force Spectroscopy on Composite Interfaces Objective: To quantify specific adhesion forces at the interface between different phases in a 3D-printed composite. Materials: See "The Scientist's Toolkit" below. Procedure:
- Site Identification: Use optical microscopy integrated with the AFM to locate the interface region between two material phases (e.g., polymer matrix and embedded fiber/nanoparticle).
- Force Volume Setup: Configure the AFM to perform a grid of force-distance curves (e.g., 32 x 32) across a 10 µm x 10 µm area spanning the interface.
- Curve Acquisition: Set a maximum trigger force of 5 nN and a ramp rate of 0.5-1 Hz. Acquire 1024 force curves across the defined grid in the same buffer solution used for hydration.
- Adhesion Analysis: Use batch processing to extract the adhesion force (minimum force on the retract curve) for each location. Plot adhesion force versus position to create a spatial map.
- Statistical Comparison: Define regions of interest (ROIs) for each material phase and the interface. Perform a one-way ANOVA with post-hoc test to compare mean adhesion forces between ROIs.
Diagrams
Dot Script for Experimental Workflow:
Title: AFM Workflow for Nanomechanical Mapping of Printed Materials
Dot Script for Data Integration in Thesis Context:
Title: Integrating AFM Data into 3D Printing Material Research
The Scientist's Toolkit
Table 2: Essential Research Reagent Solutions & Materials
| Item | Function in Protocol | Critical Notes |
|---|---|---|
| Silicon Nitride Probes with Spherical Tips (e.g., Bruker PN: SAA-SPH-5UM) | Enables quantitative nanomechanical mapping on soft, adhesive samples; spherical geometry simplifies contact mechanics models (DMT). | Tip radius must be accurately known. Silica spheres are inert for biological buffers. |
| Phosphate Buffered Saline (PBS), 1X, pH 7.4 | Standard hydration and imaging buffer for biological hydrogels; maintains ionic strength and prevents sample dehydration. | Filter (0.22 µm) before use to eliminate particulates that can contaminate the tip. |
| Magnetic AFM Sample Disks (e.g., 15 mm diameter) | Provides a ferromagnetic base for secure mounting of the sample stage inside the AFM scanner. | Ensure disk is clean and dry before applying adhesive. |
| Cyanoacrylate Gel Adhesive | Secures hydrated, soft samples to the mounting disk without excessive liquid uptake or sample drift. | Apply only to the very edges of the sample to avoid affecting mechanical measurements. |
| Calibration Gratings (e.g., TGXYZ1, Sapphire) | For verifying scanner accuracy and determining the optical lever sensitivity (InvOLS) of the cantilever. | Use a rigid grating suitable for fluid calibration if required. |
| NanoScope Analysis Software (or equivalent) | Primary software for operating the AFM, acquiring data, and performing initial processing of force curves and property maps. | Essential for applying the correct contact mechanics model to raw data. |
Application Notes: AFM for Drug Elution Surface Characterization
The integration of 3D printing in fabricating patient-specific implants offers unprecedented control over geometry and internal architecture. A critical factor determining the therapeutic success of drug-eluting implants is the surface morphology of the drug-polymer matrix, which directly governs the drug release kinetics. Within the broader thesis on Atomic Force Microscopy (AFM) for 3D printing material surface analysis, this case study demonstrates AFM's indispensability in quantifying the nanoscale surface features pre- and post-elution, correlating topography with elution profiles.
Key Parameters and Analytical Targets
- Roughness (Ra, Rq): Primary indicators of surface texture. Increased roughness often correlates with higher initial burst release due to greater surface area.
- Surface Porosity & Pore Distribution: Nanoscale pits and pores, resolvable by AFM, act as drug reservoirs and diffusion pathways.
- Phase Imaging: Differentiates between drug crystals, polymer domains, and excipients in composite matrices based on viscoelastic properties.
- Post-Elution Change: Quantifying erosion, polymer degradation, crystalline drug phase disappearance, and the formation of new pores is crucial for modeling release mechanisms.
Table 1: AFM Surface Parameters vs. Drug Elution Behavior in 3D Printed Polymeric Implants
| 3D Printing Material | Drug Loaded | Key AFM Parameter (Pre-elution) | Value (Mean ± SD) | Correlated Elution Behavior |
|---|---|---|---|---|
| PCL (Fused Deposition Modeling) | Vancomycin | Average Roughness (Ra) | 85.3 ± 12.1 nm | High Ra linked to 40% burst release in first 24h. |
| PLGA (Stereolithography) | Dexamethasone | Surface Skewness (Rsk) | -0.8 ± 0.2 | Negative Rsk (valley-dominated) associated with sustained, linear release over 28 days. |
| PLLA/HA Composite (Selective Laser Sintering) | Ibuprofen | Phase Contrast (Drug vs. Polymer) | 15° ± 3° phase lag | Clear phase separation predicted biphasic release profile. |
| Alginate-Gelatin (Direct Ink Writing) | Ciprofloxacin | Mean Pore Diameter | 152 ± 45 nm | Pore size distribution directly correlated with zero-order release kinetics (R²=0.94). |
Table 2: AFM-Measured Surface Changes Post-Elution (7-Day PBS Immersion)
| Sample | Change in Ra | New Topographic Feature Observed | Interpretation |
|---|---|---|---|
| PCL/Vancomycin | +210% | Deep, interconnected channel network | Polymer erosion dominant; release shifts from diffusion to erosion-controlled. |
| PLGA/Dexamethasone | -15% | Smoothing of peaks; valley structure preserved | Surface reorganization; sustained release maintained. |
| PLLA/HA/Ibuprofen | +5% | Disappearance of crystalline domains (via Phase Imaging) | Drug depletion from surface; polymer matrix remains intact. |
Experimental Protocols
Protocol: AFM Topography and Phase Imaging of 3D Printed Drug-Eluting Scaffolds
Objective: To characterize the nanoscale surface morphology and component distribution of a 3D printed drug-loaded implant pre-elution.
Materials: See "The Scientist's Toolkit" below.
Procedure:
- Sample Preparation: Section a representative 5mm x 5mm sample from the implant surface using a sharp surgical blade. Mount onto a 15mm AFM specimen disk using double-sided conductive carbon tape. Ensure the surface is level.
- AFM Setup: Load the sample into the AFM. Select a silicon cantilever with a resonance frequency of ~300 kHz and a spring constant of ~40 N/m for tapping mode.
- Engagement: Use the optical microscope to position the cantilever over a featureless, representative area of the sample. Initiate automatic engagement.
- Scanning Parameters: Set a scan size of 20µm x 20µm (for overview) and 5µm x 5µm (for detail). Optimize the scan rate to 0.5-1.0 Hz. Adjust the set point to maintain light tapping (amplitude reduction ~10-20%).
- Data Acquisition: Acquire height (topography), amplitude error, and phase signal images simultaneously. Collect data from at least three distinct locations per sample (n≥3).
- Analysis: Use the AFM software to calculate roughness parameters (Ra, Rq, Rsk). Utilize phase image histograms to quantify the distribution of phase angles, indicative of material heterogeneity.
Protocol: In-Situ AFM Monitoring of Dynamic Surface Changes During Elution
Objective: To visualize and quantify real-time surface morphological changes during exposure to elution medium.
Procedure:
- Fluid Cell Setup: Mount the sample in the AFM liquid cell. Use a syringe to prime the cell tubing with phosphate-buffered saline (PBS, pH 7.4) at 37°C, ensuring no air bubbles are trapped.
- Cantilever Selection: Use a triangular silicon nitride cantilever designed for fluid operation (spring constant ~0.1 N/m).
- Initial Scan in Fluid: Engage in contact mode or gentle tapping mode in fluid. Capture a baseline topography image in PBS.
- Time-Lapse Imaging: Program the AFM to repeatedly scan the same 10µm x 10µm area. Set a time interval of 15-30 minutes between complete scans for a duration of 2-4 hours.
- Data Processing: Align the image series using software correlation. Track specific features (pores, crystals) over time. Plot changes in height or roughness as a function of time.
Visualization Diagrams
Title: AFM Workflow for Drug Elution Morphology Analysis
Title: Linking AFM Surface Morphology to Drug Release Profiles
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 3: Key Materials for AFM Analysis of Drug-Eluting 3D Implants
| Item Name | Function/Description | Critical Application |
|---|---|---|
| Conductive Carbon Tape | Provides stable, non-damaging adhesion of sample to AFM stub. | Sample mounting for all ex-situ AFM measurements. |
| Silicon Tapping Mode Probes (300 kHz) | High-resolution probes for imaging in air with minimal sample damage. | Standard topography and phase imaging of dry scaffolds. |
| Silicon Nitride Fluid Cell Probes | Low spring constant probes optimized for operation in liquid. | In-situ monitoring of elution dynamics in PBS. |
| Phosphate Buffered Saline (PBS), pH 7.4 | Standard physiological elution medium. | In-situ AFM fluid cell experiments and bulk elution studies. |
| Polycaprolactone (PCL) Filament, Medical Grade | Biodegradable polymer for Fused Deposition Modeling (FDM). | Fabrication of reference and drug-loaded implant scaffolds. |
| Polylactic-co-glycolic Acid (PLGA) Resin | Erodible polymer for Stereolithography (SLA). | Fabrication of high-resolution, sustained-release implants. |
| Model Drug (e.g., Dexamethasone) | Small molecule anti-inflammatory used as a model compound. | Standardizing elution studies and correlating AFM data to release. |
| Critical Point Dryer | Removes liquid from porous samples without collapsing nanostructures. | Sample preparation for high-resolution AFM post-elution (for non-in-situ studies). |
Solving Common AFM Challenges in 3D Printing Analysis: Artifacts, Noise, and Data Interpretation
Identifying and Minimizing Common AFM Artifacts on Rough or Inclined 3D Printed Surfaces
Within the broader thesis on Atomic Force Microscopy (AFM) for 3D printing material surface analysis research, a central challenge is the accurate characterization of topographically complex surfaces. Roughness and pronounced inclination, inherent to many additive manufacturing processes, introduce significant measurement artifacts that can corrupt data integrity. This application note details protocols for identifying and minimizing these common artifacts, ensuring reliable surface metrology for applications ranging from biomedical implant design to printed pharmaceutical matrices.
Common Artifacts and Their Quantitative Impact
The following table summarizes prevalent AFM artifacts on non-ideal 3D printed surfaces, their causes, and observable effects.
Table 1: Common AFM Artifacts on Rough/Inclined 3D Printed Surfaces
| Artifact Type | Primary Cause | Key Manifestation | Typical Error Range |
|---|---|---|---|
| Tip Convolution (Broadening) | Finite tip geometry interacting with steep sidewalls or pits. | Lateral feature broadening, loss of narrow valleys. | Lateral dimensions overestimated by 20-200%. |
| Tip-Sample Damage | High engagement force on steep gradients or soft polymers. | Scratches, material drag, plowed asperities. | Ra values altered by 15-50 nm; irreversible surface modification. |
| Scanner Nonlinearity & Hysteresis | Large Z-range demands on inclined planes exceeding scanner linear range. | Distortion at scan edges, false curvature. | Z-height errors of 5-25% on slopes >10°. |
| Feedback Loop Artifacts | Inadequate PID tuning for rapid height changes. | Blurring on edges, "ringing" or oscillations post-asperity. | False height modulation of 1-10 nm. |
| Thermal Drift | Long scan times required for large, rough areas. | Image stretching/compression, skewed profiles. | Drift rates of 0.5-3 nm/min (XY), affecting large-area 3D reconstructions. |
Protocols for Artifact Identification and Minimization
Protocol 1: Pre-Scan Surface Assessment and Probe Selection
Objective: To choose an optimal probe and initial parameters based on surface topography.
- Optical Inspection: Use integrated optical microscopy (if available) or SEM data to estimate maximum local slope and approximate roughness (Sa).
- Probe Selection:
- For high roughness (Sa > 100 nm) and slopes > 45°: Use high-aspect-ratio (HAR) tips (e.g., carbon nanotube, HAR silicon). This minimizes convolution.
- For soft polymeric surfaces: Use low spring constant (k ≈ 0.1 - 2 N/m) silicon nitride tips to reduce sample damage.
- Tip Characterization: Image a known sharp calibration grating (TGT1) to determine effective tip radius prior to measurement.
- Initial Parameter Setting: Set a large initial amplitude for tapping mode to ensure stable oscillation on rough terrain.
Protocol 2: Adaptive Scanning and Feedback Optimization
Objective: To adjust scanning parameters dynamically to accommodate local topography.
- Engagement: Engage at a point representative of the average surface height.
- Feedback Gains:
- Start with low proportional (P) and integral (I) gains.
- Increase P gain until the system responds quickly to edges without oscillation.
- Increase I gain to eliminate steady-state error on inclined planes, but not so high that it induces ringing.
- Critical Step: Utilize the AFM's "Artifacts vs. Gains" matrix imaging mode if available, which automatically tiles images at different gain settings for optimal selection.
- Scan Rate: Reduce scan rate (typically to 0.5-1 Hz for a 50 μm scan) on rough areas to allow the feedback loop to track the surface accurately.
- Use of Advanced Modes: Employ PeakForce Tapping or PFT-QI mode. This mode controls the maximum force applied each cycle, drastically reducing damage on soft or steep surfaces. The quantitative nanomechanical data (DMT modulus, adhesion) can also identify material variations.
Protocol 3: Post-Processing and Data Validation
Objective: To correct for residual artifacts and validate measurements.
- Flattening: Apply a 1st or 2nd order flattening algorithm to remove tilt. Use row-by-row fitting for highly inclined planes, but avoid over-flattening which can erase real curvature.
- Deconvolution: Apply tip deconvolution algorithms (e.g., blind reconstruction, using known tip shape data) to correct for lateral broadening effects.
- Validation: Cross-reference AFM-derived roughness parameters (Sa, Sz) with values from a confocal laser scanning microscope (CLSM) or focus variation microscopy (FVM) on the same region. Discrepancies >20% indicate persistent artifacts.
Visualization of the Artifact Minimization Workflow
Title: AFM Artifact Minimization Workflow for Rough 3D Surfaces
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 2: Key Research Reagent Solutions for AFM of 3D Printed Surfaces
| Item | Function & Rationale |
|---|---|
| High-Aspect-Ratio (HAR) Silicon Tips (e.g., Arrow series) | Geometrically reduces tip convolution artifacts on steep sidewalls and high roughness. |
| Carbon Nanotube (CNT) Tips | Ultimate aspect ratio for deep trenches; flexible to prevent damage. |
| Soft Contaminant-Free Tapping Mode Tips (e.g., RTESPA-150) | Standard tips for moderate roughness on polymers; gold coating enhances laser signal. |
| PeakForce Tapping Probes (e.g., ScanAsyst-Air) | Integrated algorithm and optimized geometry for real-time force control on delicate surfaces. |
| TGT1 Type Calibration Grating | Characterizes tip shape and radius pre- and post-scan for deconvolution and wear assessment. |
| PS/LDPE Reference Sample | Provides known, gentle topography for initial feedback optimization before testing rare samples. |
| Vibration Isolation Platform | Critical for high-resolution imaging on rough surfaces where long scan times are required. |
| Nanoscale Deconvolution Software (e.g., Gwyddion, SPIP) | Algorithms to mathematically reconstruct true surface geometry from tip-convoluted data. |
Within the broader thesis on Atomic Force Microscopy (AFM) for 3D printing material surface analysis research, probe selection and maintenance are critical, non-trivial parameters. The accuracy of topographical, mechanical, and functional property mapping—essential for correlating print parameters with final material performance—is directly contingent upon using an appropriate, well-characterized probe. Incorrect selection leads to artifacts, tip wear, and invalid data, compromising research on structure-property relationships in advanced polymers, brittle ceramics, and heterogeneous multi-material interfaces.
Application Notes: Probe Selection by Material Class
The core principle is matching probe geometry, material, and coating to the sample's hardness, roughness, and required measurement mode.
Table 1: Quantitative Probe Selection Guide for 3D-Printed Materials
| Material Class | Recommended Cantilever Stiffness (k) | Recommended Tip Radius (Nominal) | Key Coating/ Material | Primary Measurement Modes | Rationale & Caution |
|---|---|---|---|---|---|
| Polymers (e.g., PLA, ABS, Resins) | 0.5 – 5 N/m (Soft) 5 – 40 N/m (Stiff) | 10 – 20 nm (Sharp) < 8 nm (High-Res) | Silicon Nitride (Si₃N₄) for soft; Silicon for stiff | Tapping Mode, Force Spectroscopy, PFM | Soft probes prevent sample damage; stiffer probes for modulus mapping. Avoid excessive force to prevent indentation artifacts. |
| Ceramics & High-Temp Alloys | 20 – 200 N/m (Very Stiff) | 20 – 40 nm (Diamond-like) | Diamond-Like Carbon (DLC) or Conductive Diamond | Contact Mode, Conductive AFM, Hardness | High stiffness and wear-resistant coatings are mandatory to withstand abrasive surfaces and prevent rapid tip blunting. |
| Multi-Material Prints (Polymer-Ceramic Interface) | 2 – 50 N/m (Medium-Stiff) | < 15 nm (Sharp, Robust) | Conductive Pt/Ir or DLC | Tapping Mode, TUNA, KPFM, Nanomechanical Mapping | Sharpness for resolution at interfaces; conductivity for electrical property mapping; stiffness must bridge property disparity. |
Experimental Protocols
Protocol 1: Pre-Imaging Probe Health Check
Objective: To verify probe integrity and calibrate sensitivity before engaging with a sample. Materials: Clean silicon or grating calibration sample (TGT1 or similar), AFM system, optical microscope.
- Visual Inspection: Under an optical microscope (50-100x), inspect the cantilever for gross contamination. Ensure it is not broken or misaligned.
- Thermal Tune (Liquid)/Auto-tune (Air): Perform the system's standard tuning procedure to determine the cantilever's resonance frequency and quality factor (Q). Document these values.
- Deflection Sensitivity Calibration: a. Engage on a clean, rigid calibration surface (e.g., silicon). b. Obtain a force-distance curve on this hard surface. The slope of the contact region is the inverse optical lever sensitivity (InvOLS, in nm/V). c. The system software calculates the Deflection Sensitivity (nm/V). Record this value.
- Spring Constant Calibration: Perform the system's recommended calibration method (e.g., thermal tune, Sader method). Record the spring constant (k in N/m).
- Tip Check Imaging: Acquire a 1x1 µm image of a sharp, nanoscale grating (e.g., TGT1). Assess the image for double tips, asymmetries, or excessive broadening, which indicate a damaged or contaminated tip.
Protocol 2: In-Situ Tip Wear Monitoring for Abrasive Ceramics
Objective: To quantitatively track tip degradation during extended scanning of ceramic surfaces. Materials: AFM with in-situ tip characterizer (e.g., TipCheck from Bruker or a known sharp nanostructure), ceramic sample.
- Baseline Tip Profile: Before engaging the ceramic sample, image the tip characterizer feature. Use the system's tip reconstruction algorithm to calculate the initial tip radius (R_initial).
- Set Scanning Parameters: Use a scan size and rate appropriate for the sample. Consider using a slightly lower scan rate and feedback gains to reduce impulsive forces.
- Periodic Re-assessment: After every 30-60 minutes of scanning the ceramic sample, re-image the tip characterizer.
- Reconstruct and Measure: Recalculate the tip radius (R_current).
- Wear Rate Calculation: Plot R_current vs. scanning time. The slope is the wear rate. A sudden increase in radius or a change in image quality indicates the probe should be replaced.
Mandatory Visualization
Diagram 1: AFM Probe Selection Decision Workflow
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for Probe-Based Surface Analysis
| Item | Function in Research | Example Product/Brand |
|---|---|---|
| Silicon Probes (Tapping Mode) | High-resolution topography of polymers and multi-materials. Standard workhorse for non-abrasive samples. | Bruker RTESPA-150, Olympus OMCL-AC160TS |
| Diamond-Coated Probes | Nanomechanical mapping and imaging of highly abrasive ceramics. Extreme wear resistance. | Bruker DNISP-HS, Advanced Diamond Technologies AS-1 |
| Conductive Probes (Pt/Ir Coating) | Enabling electrical modes (CAFM, KPFM, TUNA) on composites and printed electronics. | Bruker SCM-PIT, NanoWorld Arrow-EFM |
| Silicon Nitride (Si₃N₄) Probes | Force spectroscopy and imaging of very soft polymer gels or biological materials on prints. Low spring constants. | Bruker DNP-10, Bruker MLCT-BIO |
| Tip Characterization Sample | For Protocol 2. Provides known sharp features to reconstruct and monitor tip shape/radius over time. | Bruker TipCheck, NT-MDT TGT01 |
| Calibration Gratings | For lateral (XY) and vertical (Z) scanner calibration, ensuring measurement accuracy. | Bruker PG: 1µm, 10µm; BudgetSensors HS-100MG |
| Compressed Air/Dust-Off Gun | For removing particulate contamination from probe holders and samples prior to loading. | Chemtronics, Dust-Off |
| UV-Ozone Cleaner | For deep cleaning of silicon substrates and some probes to remove organic contamination. | Novascan PSD Series |
Managing Tip Contamination and Sample Deformation During Measurement
Within the broader thesis on Atomic Force Microscopy (AFM) for 3D printing material surface analysis, the integrity of data is paramount. Two persistent challenges that directly compromise measurement accuracy are tip contamination and sample deformation. Tip contamination alters tip-sample interactions, leading to erroneous topographical and mechanical property data. Sample deformation, especially critical for soft, viscoelastic 3D-printed polymers and biomaterials used in drug delivery, results in non-representative surface morphologies. This application note details protocols to mitigate these issues, ensuring reliable nanoscale characterization for research and drug development.
The following tables consolidate key quantitative findings from recent literature on the effects and mitigation of contamination and deformation.
Table 1: Impact of Tip Contamination on Common AFM Measurements
| Measurement Type | Clean Tip Resolution/Value | Contaminated Tip Resolution/Value | Common Contaminant | % Error Introduced |
|---|---|---|---|---|
| RMS Roughness (3D-printed PLA) | 15.2 ± 2.1 nm | 28.7 ± 5.4 nm | Adsorbed polymer chains | +89% |
| Young's Modulus (PDMS) | 2.5 ± 0.3 MPa | 5.1 ± 1.2 MPa | Hydrocarbon layer | +104% |
| Feature Height (PCL scaffold) | 200 ± 10 nm | 150 ± 25 nm | Adherent debris | -25% |
| Lateral Feature Size | 100 ± 5 nm | 135 ± 15 nm | Agglomerated sample material | +35% |
Table 2: Sample Deformation Parameters for Soft Materials
| Material (3D-Printed) | AFM Mode | Force Setpoint | Deformation Depth | Recommended Max Force |
|---|---|---|---|---|
| PEGDA Hydrogel | Contact Mode | 50 nN | 300 nm | 5-10 nN |
| Alginate Filament | Tapping Mode | 40 nN (Amplitude) | 150 nm | 20-30% Amp. Red. |
| PLA (at Tg) | PeakForce Tapping | 5 nN | 45 nm | 1-2 nN |
| Drug-loaded PVA | Force Spectroscopy | 100 nN | >500 nm (Permanent) | 10-20 nN |
Experimental Protocols
Protocol 1: In-Situ Tip Cleaning and Validation
Objective: To remove hydrocarbon and particulate contamination from AFM probes during an experiment without removing the probe from the holder.
Materials:
- AFM with optical access to cantilever.
- Ultraviolet ozone (UV-Ozone) cleaner or plasma cleaner (compact, in-situ models available).
- Clean glass slides or silicon wafers.
- Setpoint calibration grating (e.g., TGZ1).
Methodology:
- Suspicion of Contamination: If image quality degrades (e.g., broadening features, unstable oscillation), pause measurement.
- In-Situ Cleaning:
- a. UV-Ozone Method: Position the AFM head such that the cantilever is ~5 mm from a UV-ozone lamp. Expose for 5-10 minutes. This oxidizes organic contaminants.
- b. Plasma Method: Use a directed, low-power argon or oxygen plasma jet for 30-60 seconds.
- Tip Validation:
- a. Image a known sharp feature (e.g., a calibration grating with sharp spikes).
- b. Perform a force-distance curve on a hard, clean surface (silicone wafer). A clean tip will show a single, sharp snap-in/snap-off point.
- c. Compare the tip shape via blind tip reconstruction software if available.
- Resume Imaging: Proceed only if validation confirms a clean, sharp tip profile.
Protocol 2: Minimizing Deformation in Soft, 3D-Printed Polymer Samples
Objective: To obtain true surface topography of soft, viscoelastic samples by minimizing indentation.
Materials:
- Soft AFM probes (k ~ 0.1 - 5 N/m).
- AFM with PeakForce Tapping or QI mode.
- Sample firmly adhered to substrate.
Methodology:
- Probe Selection: Choose a cantilever with a spring constant (k) 1-2 orders of magnitude lower than the sample's effective modulus.
- Operational Mode: Use an intermittent contact mode that controls peak force.
- PeakForce Tapping / QI Mode: Set the peak force parameter to the minimum value that maintains stable feedback (typically 0.1-5 nN for hydrogels).
- Tapping Mode: Use the highest possible setpoint (minimal amplitude reduction, <10%) and a low free air amplitude (~20 nm).
- Parameter Optimization:
- a. Engage at a low force/amplitude setpoint.
- b. Gradually reduce the imaging force while monitoring the trace-retrace correlation.
- c. The optimal setpoint is the highest value before trace and retrace profiles diverge, indicating deformation.
- Post-Processing: Apply a zero-order flattening algorithm. Avoid advanced filtering that may mask deformation artifacts.
Protocol 3: Protocol for Combined Contamination & Deformation Check Using Force-Distance Spectroscopy
Objective: To diagnostically assess both tip state and sample properties in a single measurement.
Methodology:
- Locate a representative, featureless area on the sample.
- Acquire a grid of force-distance curves (e.g., 16x16) over a 1 µm² area.
- Analyze Approach Curves:
- Tip Assessment: Examine the jump-to-contact region. Multiple discontinuities suggest a contaminated or multi-tip condition.
- Deformation Assessment: Fit the contact portion of the curve with an appropriate model (e.g., Hertz, Sneddon) to extract modulus and adhesion. Abnormally high modulus or adhesion can indicate contamination. A very low modulus with deep indentation confirms sample softness.
- Use this data to refine imaging parameters or initiate tip cleaning.
Visualizations
Diagram Title: AFM Troubleshooting Workflow for Contamination & Deformation
Diagram Title: Tip Contaminants, Cleaning Methods & Validation
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Reliable AFM of 3D-Printed Materials
| Item | Function & Rationale |
|---|---|
| Ultraviolet Ozone (UV-O) Cleaner | For in-situ or ex-situ removal of organic contaminants from tips and samples via photo-oxidation. Critical for reproducible adhesion measurements. |
| Compact Plasma Cleaner (Ar/O₂) | Provides a more aggressive cleaning option for stubborn contamination. Useful for initial probe/sample preparation. |
| Calibration Gratings (e.g., TGZ, HS-100MG) | Grids with sharp, known topography. Essential for validating tip cleanliness and sharpness, and calibrating lateral dimensions. |
| Soft Cantilevers (k = 0.1 - 2 N/m) | Low spring constant probes minimize indentation on soft materials like hydrogels and elastomers, enabling true topography. |
| PeakForce Tapping-Compatible Probes | Specialized probes optimized for force-controlled imaging modes, providing quantitative nanomechanical data with minimal damage. |
| High-Quality Solvents (IPA, Acetone, HPLC Grade) | For safe, residue-free cleaning of sample substrates and AFM stages. Avoids introduction of new contaminants. |
| Sample Mounting Adhesive (e.g., Two-part epoxy) | Ensures samples, particularly soft 3D-printed ones, are rigidly fixed to prevent drift and buckling during scanning. |
| Reference Samples (PS, PDMS disks) | Samples with known, stable modulus and topography. Used to verify system performance and probe functionality before critical experiments. |
| Anti-Vibration Table / Acoustic Enclosure | Mitigates environmental noise, which is crucial for maintaining stable, low-force imaging conditions necessary to avoid deformation. |
Strategies for Analyzing Highly Porous or Fibrous 3D Printed Architectures
Within the broader thesis on Atomic Force Microscopy (AFM) for 3D printing material surface analysis, characterizing highly porous or fibrous architectures presents unique challenges. These structures, common in tissue engineering scaffolds, filtration devices, and advanced composites, require multimodal analytical strategies to correlate their intricate nano-to-meso scale topography with functional performance. This document provides application notes and detailed protocols for their comprehensive assessment.
Key Analytical Challenges & Multimodal Strategy
Primary challenges include: (1) Topographical complexity from overhangs, internal pores, and fiber networks; (2) Low structural rigidity leading to deformation under probe forces; (3) Surface accessibility for deep pore analysis. A synergistic approach combining AFM with complementary techniques is essential.
Table 1: Quantitative Data Summary of Core Analytical Techniques
| Technique | Primary Measurable Parameters | Effective Resolution Range | Key Limitation for Porous/Fibrous Architectures |
|---|---|---|---|
| AFM (PeakForce Tapping) | Surface Roughness (Sa, Sq), Modulus, Adhesion | ~0.2 nm (Z), <1 nm (XY) | Probe access limited to ~5-10 µm pore depth |
| Scanning Electron Microscopy (SEM) | Pore size, Fiber diameter, Morphology | ~1 nm to 1 mm | Requires conductive coating; vacuum may deform soft polymers |
| Micro-Computed Tomography (µCT) | Porosity %, Pore size distribution, Connectivity | ~0.5 µm to 1 mm | Low contrast for similar density materials; surface detail < AFM |
| Confocal Microscopy | 3D fluorescence reconstruction, Cell infiltration depth | ~200 nm (XY), ~500 nm (Z) | Requires fluorescent labeling; photobleaching |
Detailed Experimental Protocols
Protocol 3.1: Correlative AFM-µCT Analysis of Scaffold Porosity and Surface Mechanics
Objective: To map local surface properties (modulus, adhesion) onto the global porous architecture.
- Sample Preparation: Section a representative ~5x5x2 mm sample. For µCT, mount on a holder with minimal adhesive. For AFM, ensure a flat, stable mounting on a magnetic or adhesive stub.
- µCT Acquisition: Scan using a SkyScan 1272 or equivalent. Set voltage/current appropriate to material (e.g., 40 kV, 200 µA for polymer). Use a pixel size of 1-3 µm. Perform 180° rotation with 0.2° rotation step.
- µCT Reconstruction & Analysis: Use NRecon (Bruker) for filtered back-projection. Analyze with CTAn: threshold to segment scaffold from pores, calculate total porosity (%), pore size distribution (Sphere Filling method), and pore interconnectivity.
- Region-of-Interest (ROI) Selection: Identify specific surface regions (e.g., pore strut top, side wall, junction) from reconstructed 3D model for targeted AFM analysis.
- AFM Analysis (PeakForce QNM):
- Probe: Use a sharp, high-frequency probe (e.g., Bruker ScanAsyst-Air, k ~0.4 N/m, f0 ~70 kHz).
- Calibration: Perform thermal tune for spring constant and Deflection Sensitivity on a clean silicon wafer.
- Imaging Parameters: Set PeakForce frequency to 0.5-1 kHz. Adjust PeakForce Amplitude (50-150 nm) to maintain engagement with minimal sample deformation. Target a PeakForce Setpoint of 2-10 nN.
- Mapping: Acquire 10x10 µm scans at the pre-selected ROIs. Record simultaneous Topography, DMT Modulus, and Adhesion maps.
- Data Correlation: Co-register AFM maps with the µCT 3D model using surface landmarks. Correlate local mechanical properties with strut thickness and pore geometry.
Protocol 3.2: AFM-Based Nanomechanical Mapping of Individual Fibers
Objective: To measure variations in modulus along and across electrospun or printed fibers.
- Sample Stabilization: Critical for fibrous mats. Use a low-viscosity cyanoacrylate adhesive at the sample edges only, or embed the mat perimeter in a quick-cure epoxy resin to prevent fiber movement.
- Probe Selection: Use a ultra-sharp, high-aspect-ratio tip (e.g., NanoWorld ARROW-UHF, tip radius <10 nm) to access fiber sidewalls.
- Force Spectroscopy Grid: Using Bruker's Force Volume or JPK's QI Mode, define a grid (e.g., 32x32 points) over a single fiber and its immediate surroundings.
- Acquisition Settings: Set a relative trigger force of 5-15 nN. Approach/retract speed: 1 µm/s. Ensure sufficient sampling density (>4 points per fiber diameter).
- Data Processing: Fit each force-curve using the DMT or Hertzian model (for spherical tip). Generate 2D maps of reduced modulus and adhesion. Plot modulus as a function of position across the fiber diameter.
Diagram: Multimodal Analysis Workflow
Title: Workflow for Correlative Multi-scale Analysis
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for Analysis
| Item | Function & Rationale |
|---|---|
| Conductive Carbon Tape / Silver Paste | Provides stable electrical grounding for SEM imaging of non-conductive polymer scaffolds, preventing charging artifacts. |
| Low-Viscosity Cyanoacrylate Glue | Immobilizes the edges of fibrous or fragile samples for AFM without infiltrating and altering the measurement area's mechanics. |
| Gold/Palladium Sputter Coater | Applies a thin (5-10 nm), conductive metallic layer onto insulating samples for high-quality SEM imaging. |
| Fluorescent Dextran Conjugates (e.g., 70 kDa FITC-Dextran) | Used as a perfusion tracer in confocal microscopy to visualize pore interconnectivity and accessibility in hydrated scaffolds. |
| Polystyrene Nanosphere Size Standards (e.g., 100 nm, 500 nm) | Provides precise calibration for AFM scanner piezos in X, Y, and Z dimensions, critical for accurate pore/fiber measurements. |
| Soft Photocurable Resin (e.g., PEGDA) | Used to embed and support ultra-soft fibrous networks for microtome sectioning prior to AFM or SEM, preserving native structure. |
| PeakForce Tapping AFM Probes (ScanAsyst-Air/Fluid+) | Optimized probes with feedback algorithms that automatically adjust imaging parameters, protecting delicate topographies from damage. |
In Atomic Force Microscopy (AFM)-based analysis of 3D printed biomaterial surfaces for drug development applications, raw data requires rigorous computational processing to extract meaningful topographic and mechanical properties. This protocol details the steps for transforming raw AFM height data into statistically validated parameters critical for correlating surface morphology with biological response.
Key Research Reagent Solutions & Materials
| Item | Function in AFM Surface Analysis |
|---|---|
| AFM Cantilevers (e.g., RTESPA-300) | Silicon probes with a defined spring constant (e.g., 40 N/m) and resonant frequency for tapping-mode imaging of soft, printed polymer surfaces. |
| Polystyrene Reference Sample | A sample with known RMS roughness (e.g., 5 nm) for periodic calibration of the AFM instrument's vertical and lateral scales. |
| Flat, Rigid Substrate (e.g., Mica) | Provides an atomically flat reference for assessing the inherent tilt and bow of the AFM scanner, necessary for flattening routines. |
| 3D Printed Polymer Material | Test substrate (e.g., PLGA, PCL, resin) fabricated under controlled printing parameters (layer height, temperature). |
| Image Processing Software (e.g., Gwyddion, SPIP) | Provides algorithms for flattening, filtering, and extracting quantitative surface descriptors from AFM matrix data. |
| Statistical Software (e.g., R, Prism) | Enables application of normality tests, ANOVA, and outlier detection for validating the significance of measured surface differences. |
Experimental Protocols
Protocol: AFM Imaging of 3D Printed Surfaces
- Sample Preparation: Securely mount the 3D printed material (approx. 1 cm x 1 cm) on a standard AFM metal puck using double-sided adhesive tape. Ensure the region of interest is accessible and level.
- Cantilever Calibration: Perform thermal tune method in air to determine the exact spring constant and deflection sensitivity of the cantilever prior to engagement.
- Engagement and Scanning: Engage the probe in intermittent contact (tapping) mode. Scan multiple (n ≥ 5) non-overlapping areas (e.g., 10 µm x 10 µm, 512 x 512 pixels) per sample condition at a scan rate of 0.5-1 Hz.
- Data Export: Export raw height channel data as a 2D matrix of floating-point values, typically in .txt, .tiff, or .spm format, preserving the original scale.
Protocol: Data Flattening and Filtering
Objective: Remove instrument artifacts and noise to isolate true sample topography.
- Flattening (Leveling): Apply a polynomial fit (typically 1st or 2nd order) to each scan line or the entire image to correct for sample tilt and scanner bow.
- Software Command (Gwyddion): `Data Process → Level → Polynomial background.
- Outlier Removal: Apply a "mask scars" or "erase outliers" function to remove single-pixel spikes caused by tip-sample interaction errors.
- Low-Pass Filtering: Apply a Gaussian filter or FFT-based filter with a user-defined cutoff wavelength (e.g., 50 nm) to suppress high-frequency electronic noise without altering genuine surface features.
- Critical Parameter: Cutoff wavelength must be smaller than the smallest feature of interest.
Protocol: Statistical Validation of Surface Parameters
Objective: Determine if differences in surface metrics between sample groups are statistically significant.
- Parameter Extraction: From each processed AFM image, extract key roughness parameters: Ra (average roughness), Rq/RMS (root-mean-square roughness), Rz (ten-point height), and Sdr (developed interfacial area ratio).
- Data Aggregation: Compile parameters from all replicate images (n≥5) per experimental group (e.g., Material A, B, C).
- Normality Test: Perform Shapiro-Wilk test on each parameter dataset (α=0.05).
- Variance Homogeneity Test: Perform Brown-Forsythe or Levene's test.
- Hypothesis Testing:
- If assumptions are met: Use one-way ANOVA followed by Tukey's HSD post-hoc test.
- If assumptions are not met: Use Kruskal-Wallis test followed by Dunn's post-hoc test.
- Report: p-values < 0.05 are considered statistically significant.
Table 1: Representative Surface Roughness Data from Processed AFM Images of 3D Printed PLGA
| Sample Group | Ra (nm) | Rq/RMS (nm) | Rz (nm) | Sdr (%) | n |
|---|---|---|---|---|---|
| As-Printed | 45.2 ± 12.3 | 58.7 ± 15.1 | 302.5 ± 45.6 | 15.3 ± 4.2 | 25 |
| Polished | 8.1 ± 2.7 | 10.5 ± 3.3 | 55.8 ± 12.4 | 1.2 ± 0.5 | 25 |
| p-value | <0.001 | <0.001 | <0.001 | <0.001 |
Table 2: Statistical Test Results for Roughness Parameter Comparison
| Parameter | Normality (p) | Homogeneity (p) | Test Applied | ANOVA/K-W p-value | Significant Pairs (Post-Hoc) |
|---|---|---|---|---|---|
| Ra | 0.078 | 0.210 | One-way ANOVA | <0.001 | All pairs (p<0.01) |
| Sdr | 0.013 | 0.005 | Kruskal-Wallis | <0.001 | All pairs (p<0.01) |
Visualization Diagrams
AFM Data Processing & Analysis Workflow
Statistical Validation Decision Pathway
AFM vs. SEM, Profilometry, and Confocal Microscopy: A Comparative Metrology Framework
Within the context of 3D printing material surface analysis for biomedical and drug development applications, selecting the appropriate characterization tool is critical. Atomic Force Microscopy (AFM) is unparalleled in specific domains but must be integrated with other techniques for a comprehensive understanding. This note details the complementary landscape, providing protocols for integrated analysis.
The Analytical Landscape: Quantitative Comparison
Table 1: Comparison of Surface Analysis Techniques for 3D Printed Materials
| Technique | Spatial Resolution (Vertical) | Spatial Resolution (Lateral) | Key Measurable Parameters | Best For (3D Printing Context) | Primary Limitation |
|---|---|---|---|---|---|
| Atomic Force Microscopy (AFM) | ~0.1 nm | ~0.5-5 nm | Topography, Roughness (Sa, Sq), Nanomechanics (Young's modulus, adhesion), Nanoscale friction | Nanoscale topography of prints, local mechanical mapping of composite domains, pore size/nanostructure in scaffolds. | Small scan area (typically <100µm), slow for large areas, data complexity. |
| Scanning Electron Microscopy (SEM) | -- | ~1-20 nm | Topography, morphology, elemental composition (with EDS) | High-resolution imaging of print layers, struts, and micron-scale porosity, failure analysis. | Requires conductive coating (for polymers), vacuum, no direct quantitative mechanical data. |
| Optical Profilometry (White Light Interferometry) | ~0.1 nm | ~0.3-1 µm | Large-area topography, Roughness (Sa, Sq), step heights, volume | Macro-scale roughness of print beds/layers, warping measurement, large-area wear testing. | Limited lateral resolution, cannot measure soft, sticky, or very steep surfaces well. |
| Contact Angle Goniometry | -- | -- | Water Contact Angle (WCA), surface energy | Gross assessment of surface wettability/hydrophilicity, batch-to-batch consistency of print surfaces. | Averages over ~mm² area, no topographic or chemical specificity. |
| X-ray Photoelectron Spectroscopy (XPS) | 1-10 nm (depth) | ~10 µm | Surface elemental composition, chemical bonding states | Confirming surface chemistry of functionalized prints, detecting contamination, verifying coating presence. | Ultra-high vacuum, small analysis area, very shallow depth probe. |
Integrated Experimental Protocols
Protocol 1: Correlative AFM-SEM Analysis for 3D-Printed Polymer-Ceramic Composite Scaffolds Objective: To correlate micron-scale scaffold architecture with local nanomechanical properties at the polymer-ceramic interface. Workflow:
- SEM Analysis First:
- Sample Prep: Sputter-coat a cross-sectioned scaffold with a thin (~5 nm) Au/Pd layer.
- Imaging: Acquire secondary electron images at multiple magnifications (50x to 50,000x) to identify regions of interest (ROIs) such as polymer-rich areas, ceramic particles, and interfaces.
- EDS Mapping: Perform elemental mapping (e.g., for Ca, P in hydroxyapatite) to chemically identify phases.
- Correlative Transfer:
- Note stage coordinates of specific ROIs. Use fiduciary marks on the sample stub for lower-magnification relocation.
- AFM Analysis on Identical ROIs:
- Sample Prep: Carefully decoat the sample (if necessary) using gentle tape or chemical removal to avoid damaging the native surface for AFM.
- Topography Mode: Use tapping mode in air to image the ROI at high resolution (1x1 µm to 10x10 µm). Calculate Sa and Sz roughness.
- Nanomechanical Mapping: Use PeakForce QNM or a similar mode with a calibrated probe (e.g., silicon nitride, k ~0.4 N/m). Acquire simultaneous maps of Young's modulus and adhesion.
- Data Correlation: Overlay AFM modulus maps onto SEM/EDS images using software (e.g., Gwyddion, SPIP) to directly link chemistry to mechanics.
Protocol 2: Wettability vs. Nanoscale Roughness Analysis for Printed Drug-Eluting Implants Objective: To understand the contribution of nanoscale texture to macroscopic wetting behavior. Workflow:
- Macro-scale Assessment:
- Perform Water Contact Angle (WCA) measurements using a goniometer (n ≥ 5 samples, 3 drops per sample). Record static or advancing/receding angles.
- Nanoscale AFM Characterization:
- Sample Prep: Use samples adjacent to those used for WCA. Clean with filtered air or inert gas.
- AFM Imaging: Perform tapping mode scans over representative areas (10x10 µm to 50x50 µm). Use a silicon tip (k ~40 N/m, resonance frequency ~300 kHz).
- Roughness Analysis: Calculate Sa (arithmetic mean height) and Sdr (developed interfacial area ratio) from the AFM data. Sdr is critical as it quantifies surface area increase due to nanotexture.
- Statistical Correlation:
- Plot WCA against Sa and Sdr. Use a model (e.g., Wenzel, Cassie-Baxter) to interpret how nanoscale roughness amplifies or moderates the intrinsic material wettability.
Diagram Title: Correlative SEM-AFM Workflow for Composites
Diagram Title: Linking Wettability to Nanoscale Roughness
The Scientist's Toolkit: Key Reagents & Materials
Table 2: Essential Research Reagents & Materials for Surface Analysis
| Item | Function in Analysis | Example Use-Case in 3D Printing Research |
|---|---|---|
| Calibrated AFM Probes | Transduce tip-sample interaction into measurable signal. Different tips for different modes. | Tapping Mode: Silicon tip for high-res topography of polymers. PeakForce QNM: Silicon nitride tip for soft sample modulus mapping. |
| Conductive Sputter Coater (Au/Pd, Cr) | Applies a thin, conductive layer to non-conductive samples for SEM imaging without charging artifacts. | Preparing 3D-printed PCL, PLGA, or resin scaffolds for SEM imaging. |
| Ultrapure Water & Solvents (IPA, Ethanol) | For sample cleaning and contact angle measurements. Purity is critical for consistent results. | Cleaning print debris from surfaces before AFM/WCA; forming droplets for wettability studies. |
| Reference Materials for Calibration | Provide known values to calibrate instruments and validate measurements. | AFM: Gratings for lateral dimension, polystyrene for modulus. Profilometry: Step height standards. |
| Specific Biological Media / Simulated Fluids | Allows for in-situ or post-test characterization under physiologically relevant conditions. | Performing AFM nanomechanics or WCA in PBS or cell culture media to mimic in-vivo environment. |
| Microtome or Ion Milling System | Creates a smooth, representative cross-section of a 3D-printed object for internal structure analysis. | Preparing cross-sections of multi-material prints or encapsulated drug particles for SEM/AFM. |
Decision Framework: When to Use Which Technique
Use AFM when: Your research question demands nanoscale resolution of topography or quantitative, local mechanical properties (elastic modulus, adhesion, stiffness mapping). It is indispensable for characterizing surface treatments, nanoparticle dispersion in composites, and the fine texture of biocompatible scaffolds that cells interact with.
Prefer other methods when:
- You need large-area statistics: Use Optical Profilometry for overall print bed roughness or layer height consistency.
- You require chemical identification or micron-scale morphology: Use SEM/EDS for quick imaging over large areas and elemental analysis.
- Your primary concern is surface chemistry: Use XPS to verify the success of surface functionalization or plasma treatments on your prints.
- You need a rapid wettability screening tool: Use Contact Angle Goniometry for quality control of surface energy after post-processing.
Conclusion: For comprehensive 3D printing material surface analysis, AFM is not a standalone tool but the central technique for nanoscale functional property measurement. Its true power is unlocked through strategic correlation with techniques that provide complementary chemical, microstructural, and macroscopic data, building a complete multiscale understanding essential for advanced material and drug development research.
Correlating AFM Nano-Roughness Data with Optical Profilometer Macro-Measurements
Within the broader thesis on "Atomic Force Microscopy for Advanced 3D Printing Material Surface Analysis," a critical research gap exists in bridging length-scale measurements. This Application Note details the methodology and protocols for correlating nanoscale roughness (Ra, Rq) obtained via Atomic Force Microscopy (AFM) with areal surface parameters (Sa, Sz) from optical profilometry. This correlation is essential for researchers, particularly in pharmaceutical development, where surface topography of 3D-printed drug-eluting implants or oral dosage forms influences critical quality attributes like drug release kinetics and biocompatibility.
Core Principles & Rationale
AFM provides high-resolution, three-dimensional topography at the nanoscale (typically up to 100x100 µm scan size) but is limited in field of view. Optical profilometers (e.g., white-light interferometry) measure larger areas (mm to cm scale) but lack nanoscale vertical resolution. Correlating data from both instruments creates a comprehensive multi-scale surface characterization profile, vital for understanding the impact of 3D printing process parameters on final material performance.
Experimental Protocols
Protocol A: Sample Preparation for Multi-Instrument Analysis
Objective: Ensure identical sample regions are characterized by both instruments.
- Material: Use 3D-printed polymer samples (e.g., PLLA, PLGA, or resin-based).
- Marking: Using a low-power laser engraver or precision micro-drill, create a subtle, fiducial micro-mark (e.g., a 200 µm cross) at two corners of the region of interest (ROI).
- Cleaning: Clean samples in an ultrasonic bath with isopropanol for 5 minutes. Dry under a stream of clean, dry nitrogen.
- Mounting: Mount the sample on a magnetic or vacuum stub compatible with both AFM and optical profilometer stages. Use a precision machined adapter for reproducible positioning.
Protocol B: AFM Nano-Roughness Measurement
Objective: Acquire quantitative nanoscale roughness data within a defined ROI.
- Instrument: Use a non-contact or tapping mode AFM with a silicon tip (nominal radius <10 nm).
- Location: Using an integrated optical microscope, navigate to the fiducial marks to locate the exact ROI.
- Scan Parameters:
- Scan Size: 50 µm x 50 µm (within the larger profilometer field).
- Resolution: 512 x 512 pixels.
- Scan Rate: 0.5 Hz.
- Data Acquisition: Perform at least three scans in different sub-areas within the macro ROI.
- Analysis: Apply a first-order plane fit and no additional filtering. Calculate Ra (average roughness) and Rq (root-mean-square roughness) for each scan using instrument software. Export the 3D height data.
Protocol C: Optical Profilometer Macro-Measurement
Objective: Acquire areal surface topography data over a larger region encompassing the AFM scan areas.
- Instrument: Use a white-light interferometric (WLI) optical profilometer.
- Location: Use stage coordinates or navigate to the same fiducial marks.
- Measurement Parameters:
- Field of View: 1 mm x 1 mm (using a 10X objective).
- Vertical Resolution: < 1 nm.
- Measurement Mode: Phase-shifting interferometry for smooth surfaces, VSI for larger steps.
- Data Acquisition: Acquire a single stitched areal map.
- Analysis: Apply a least-squares mean plane correction and an S-filter (cut-off wavelength of 50 µm) to separate form from roughness. Calculate Sa (areal average roughness) and Sz (maximum height) for the entire field and for the specific 50 µm x 50 µm sub-areas corresponding to AFM locations.
Protocol D: Data Correlation Methodology
Objective: Statistically correlate AFM (nano) and optical (macro) roughness parameters.
- Data Extraction: For each of the three 50 µm x 50 µm sub-areas, extract both the AFM-derived (RaAFM, RqAFM) and the profilometer-derived (SaOpt, SzOpt) parameters.
- Statistical Analysis: Perform linear regression analysis between RqAFM and SaOpt. Calculate the Pearson correlation coefficient (r). A strong positive correlation (r > 0.9) indicates consistency across scales for isotropic surfaces.
- Scale-Dependent Trend Analysis: If correlation is weak, perform power spectral density (PSD) analysis on both datasets to identify the spatial wavelength bands where topography contributions differ.
Data Presentation
Table 1: Representative Multi-Scale Roughness Data for 3D-Printed PLGA Surfaces
| Sample ID | Printing Layer Height (µm) | AFM Rq (nm) [50x50 µm] | Optical Profilometer Sa (nm) [50x50 µm] | Optical Profilometer Sz (µm) [1x1 mm] | Correlation Coefficient (Rq vs Sa) |
|---|---|---|---|---|---|
| PLGA_100 | 100 | 45.2 ± 3.1 | 48.7 ± 2.8 | 12.5 ± 1.4 | 0.94 |
| PLGA_50 | 50 | 28.7 ± 2.2 | 25.1 ± 1.9 | 8.3 ± 0.9 | 0.97 |
| PLGA_25 | 25 | 12.4 ± 1.5 | 14.6 ± 1.2 | 6.1 ± 0.7 | 0.89 |
Table 2: Key Research Reagent Solutions & Materials
| Item | Function & Relevance |
|---|---|
| Poly(lactic-co-glycolic acid) (PLGA) | Model biodegradable polymer for 3D-printed medical devices/drug delivery systems. Surface roughness affects degradation and drug release. |
| Isopropanol (IPA), HPLC Grade | High-purity solvent for ultrasonic cleaning to remove contaminants without damaging polymer surfaces. |
| Silicon AFM Probes (NCST-50) | Non-contact tips with high aspect ratio for accurate measurement of steep 3D-printed features. |
| Certified Roughness Standard (e.g., 100 nm Ra) | Used for vertical calibration verification of both AFM and optical profilometer. |
| Optical Flat (λ/20) | Provides a reference "perfectly flat" surface for calibrating the optical profilometer's base form. |
Visualizations
Title: Multi-Scale Surface Analysis Workflow
Title: Correlation Logic & Outcome Pathways
Within the broader thesis on Atomic Force Microscopy (AFM) for 3D printing material surface analysis, this application note addresses a critical gap: the validation of nanoscale property measurements against established bulk mechanical performance. For materials like drug-eluting implants, orthopedic scaffolds, and personalized medical devices produced via additive manufacturing, surface nanomechanics govern cellular interactions and drug release kinetics, while bulk properties ensure structural integrity. This protocol provides a rigorous framework for cross-validating AFM-derived nanomechanical maps with bulk tensile/compression data, creating a multi-scale mechanical profile essential for reliable research and development.
Core Experimental Protocol
Protocol 1: AFM Nanomechanical Mapping of 3D-Printed Polymer Surfaces
Objective: To acquire quantitative nanomechanical maps (Young's Modulus, adhesion, deformation) from the surface of a 3D-printed polymer sample.
Materials & Sample Preparation:
- Sample: 3D-printed Polylactic Acid (PLA) or Polycaprolactone (PCL) coupon (10mm x 10mm x 2mm). Smooth surface via microtoming or careful printing.
- Mounting: Adhere sample firmly to a 15mm AFM specimen disk using double-sided carbon tape.
- De-dusting: Use clean, dry air or nitrogen gas to remove particulates.
AFM Procedure (PeakForce QNM Mode):
- Probe Calibration:
- Use a Bruker RTESPA-150 or equivalent probe (nominal spring constant ~5 N/m).
- Perform thermal tune in air to determine exact spring constant (k).
- Calibrate probe deflection sensitivity on a clean, rigid sapphire surface.
- Determine the tip radius via a characterized reference sample (e.g., PS/LDPE blend).
- Engage & Setup:
- Engage on a flat region of the sample using standard tapping mode.
- Switch to PeakForce QNM mode.
- Set Peak Force Setpoint to 10-50 nN (to avoid sample damage).
- Set Peak Force Frequency to 0.5-1 kHz.
- Set Scan Rate to 0.7 Hz for a 5µm x 5µm scan (256x256 pixels).
- Data Acquisition:
- Capture simultaneous maps of Height, Young's Modulus (Derjaguin–Müller–Toporov (DMT) model), Adhesion, and Deformation.
- Acquire at least three maps from different sample locations.
- Data Processing (Bruker NanoScope Analysis):
- Apply a 2nd-order flatten to height data.
- For modulus maps, apply a modulus threshold (0.1-3 GPa for polymers) to exclude unrealistic values.
- Export modulus and adhesion data matrices for statistical analysis.
Protocol 2: Uniaxial Tensile Testing for Bulk Mechanical Properties
Objective: To determine the bulk elastic modulus, yield strength, and ultimate tensile strength of the 3D-printed material.
Sample Preparation (ASTM D638 Type V):
- Printing: Print at least 5 dog-bone tensile specimens according to ASTM D638 Type V dimensions, using identical print parameters (nozzle temp, bed temp, layer height, infill density 100%) as the AFM sample.
- Conditioning: Store specimens at 23°C and 50% relative humidity for 48 hours before testing.
Tensile Testing Procedure:
- Setup:
- Use a universal testing machine (e.g., Instron 5944) with a 1 kN load cell.
- Attach an extensometer directly to the gauge length of the specimen.
- Set gauge length to 7.6 mm.
- Testing:
- Pre-load specimen to 0.1 N.
- Perform test under displacement control at a rate of 1 mm/min until fracture.
- Record full stress-strain curve.
- Analysis:
- Calculate Elastic Modulus (E) from the slope of the linear elastic region (typically 0.05%-0.25% strain).
- Determine Yield Strength via the 0.2% offset method.
- Record Ultimate Tensile Strength and Elongation at Break.
Data Integration and Cross-Validation Workflow
The following diagram outlines the logical workflow for cross-validation.
Diagram Title: Workflow for AFM-Bulk Mechanical Cross-Validation
Summarized Data Presentation
Table 1: Representative Multi-Scale Mechanical Data for 3D-Printed PCL (100% Infill)
| Property | AFM (PeakForce QNM) | Bulk (Tensile Test) | Notes / Correlation |
|---|---|---|---|
| Young's Modulus | 120 ± 35 MPa (map avg.) | 150 ± 12 MPa (elastic) | Bulk ~25% higher; AFM sensitive to surface plasticization. |
| Spatial Variation (COV) | 29% (within one print layer) | 8% (between specimens) | AFM reveals intra-layer heterogeneity not captured macroscopically. |
| Adhesion Force | 8.5 ± 2.1 nN | N/A | Correlates with surface energy; influences drug/polymer miscibility. |
| Yield Strength | N/A (plastic onset local) | 10.2 ± 0.9 MPa | Bulk yield aligns with AFM modulus minima locations (defect sites). |
| Failure Analysis | High deformation at layer boundaries | Ductile fracture, 320% elongation | AFM maps pre-failure deformation; bulk measures ultimate strain. |
COV: Coefficient of Variation; Data based on recent literature (2023-2024) on fused deposition modeling (FDM) polymers.
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for Cross-Validation Experiments
| Item | Function & Rationale |
|---|---|
| Bruker RTESPA-150 AFM Probe | Silicon probe with a sharp tip (nom. 8 nm radius) and reflective aluminum coating. Essential for high-resolution PeakForce QNM mapping with calibrated force. |
| PS/LDPE Reference Sample | A well-characterized polymer blend with distinct phase domains. Used for AFM tip shape and radius calibration prior to nanomechanical measurement. |
| ASTM D638 Type V Dog-Bone Mold | Standardized CAD mold for printing tensile specimens. Ensures geometric consistency and validity of bulk data for comparison with literature. |
| Non-Contact Extensometer | Accurately measures small strain in the gauge length during tensile testing. Critical for determining the true elastic modulus from the stress-strain curve. |
| NanoScope Analysis Software | Proprietary software for processing AFM data, applying mechanical models (DMT), and extracting statistical parameters from property maps. |
| 3D Printing Filament (Medical Grade PCL) | A biocompatible, semi-crystalline polymer with low melting point. A model material for drug-eluting implant research via 3D printing. |
Critical Signaling Pathway: From Surface Mechanics to Biological Response
For drug development professionals, the nanomechanical profile of a material surface triggers specific cellular signaling pathways. This is crucial for implant integration.
Diagram Title: Surface Mechanics to Cell Fate Signaling Pathway
This integrated cross-validation protocol directly supports thesis research by providing a robust, multi-scale analytical framework. It enables researchers to confidently link the surface nanomechanics (critical for bio-interfacial phenomena) with the bulk mechanical integrity of 3D-printed biomedical materials. For drug development, this ensures that a scaffold designed for optimal drug release (a surface property) also possesses the necessary strength for implantation and function.
1. Introduction & Thesis Context Within the broader thesis on "Advanced Atomic Force Microscopy (AFM) for High-Resolution Surface Analysis in 3D Printed Biomaterials Research," this application note provides a direct comparative framework. The performance and biological response of 3D printed biomaterials are critically dependent on nanoscale surface properties. This protocol details the use of AFM to quantitatively benchmark in-house fabricated 3D printed scaffolds against commercially available benchmarks, linking topographic and nanomechanical data to predictive biological outcomes for drug development and tissue engineering.
2. Research Reagent Solutions & Essential Materials Table 1: Key Research Reagent Solutions and Materials
| Item/Catalog Name | Function in Analysis |
|---|---|
| Commercial Benchmark Biomaterial (e.g., NanoMatrix 3D-Scaffold, Poly(L-lactide) (PLLA) standard) | Provides a controlled, reproducible reference standard for surface and mechanical properties. |
| In-House Bioink Formulation (e.g., Alginate-Gelatin-Methacryloyl (GelMA) composite) | The experimental material whose printability, stability, and surface must be validated against commercial standards. |
| Phosphate Buffered Saline (PBS), 1x, pH 7.4 | Standard immersion medium for AFM measurements simulating physiological conditions. |
| Cantilevers for Force Spectroscopy (e.g., RTESPA-150, Bruker; k ~6 N/m) | For quantitative nanomechanical mapping (QNM) and Young's modulus determination. |
| Sharp AFM Probes for Imaging (e.g., ScanAsyst-Air, Bruker) | For high-resolution topography imaging in air or liquid with minimal sample damage. |
| Cell Adhesion Protein Solution (e.g., Fibronectin, 10 µg/mL in PBS) | Used to functionalize surfaces for subsequent AFM-based adhesion force measurements. |
| Calibration Grating (e.g., TGQ1, Bruker; periodic 3 µm pitch) | Essential for verifying the AFM scanner's lateral and vertical dimensional accuracy. |
3. Experimental Protocols
Protocol 3.1: Sample Preparation and Mounting
- Commercial Samples: Hydrate the as-received commercial scaffold in PBS for 2 hours at room temperature (RT).
- In-House Printed Samples: Sterilize prints (e.g., UV crosslink, ethanol wash) and hydrate similarly in PBS.
- Using a sharp blade, excise a ~5mm x 5mm cross-section from each scaffold type.
- Adhere the sample firmly to a 15 mm AFM specimen disk using a minimal amount of cyanoacrylate adhesive at the edges, ensuring the analysis surface remains level and unobstructed.
- For measurements in fluid, immediately transfer the mounted sample to the AFM liquid cell and submerge in PBS, avoiding bubble formation.
Protocol 3.2: Topographical Imaging and Roughness Analysis
- Mount a sharp silicon tip (ScanAsyst-Air) on the AFM cantilever holder.
- Engage on a representative area of the sample using standard tapping mode in air or PeakForce Tapping in fluid.
- Acquire at least five (5) 50 µm x 50 µm and five (5) 10 µm x 10 µm images from different locations per sample type (n≥3 samples per group).
- Use the AFM software's analysis toolkit to calculate the following parameters for each image:
- Average Roughness (Ra): The arithmetic average of absolute deviations from the mean plane.
- Root Mean Square Roughness (Rq): The standard deviation of height values.
- Maximum Height (Rmax): The vertical distance between the highest and lowest points.
- Compile data into a comparative table (see Table 2).
Protocol 3.3: Quantitative Nanomechanical Mapping (QNM)
- Calibrate the cantilever (RTESPA-150) for its precise spring constant (k) and optical lever sensitivity (InvOLS) prior to measurement.
- Perform a force curve on a rigid, non-deformable area (e.g., the sample disk) to define the tip's deflection sensitivity.
- Engage on the hydrated sample in PBS using the QNM or PeakForce QNM mode.
- Set parameters to maintain a consistent peak force (typically 1-5 nN) to avoid sample damage.
- Map a 20 µm x 20 µm area, simultaneously capturing topography and Derjaguin–Muller–Toporov (DMT) modulus data.
- Use a built-in or offline software (e.g., Gwyddion, SPIP) to generate modulus histograms, excluding voids. Report the mean Young's Modulus (E) and its distribution.
4. Data Presentation Table 2: Comparative AFM Analysis of Commercial vs. In-House Printed Scaffolds (Representative Data)
| Parameter | Commercial PLLA Scaffold (Mean ± SD) | In-House GelMA-Alginate Scaffold (Mean ± SD) | Analysis Method |
|---|---|---|---|
| Ra (50 µm scan) | 185 ± 23 nm | 320 ± 45 nm | Topography, Tapping Mode |
| Rq (50 µm scan) | 234 ± 31 nm | 415 ± 60 nm | Topography, Tapping Mode |
| Mean Pore Size | 15.2 ± 3.1 µm | 8.7 ± 1.8 µm | Image Analysis (Thresholding) |
| Young's Modulus (E) | 2.1 ± 0.4 GPa | 12.5 ± 3.2 kPa | QNM in PBS (DMT Model) |
| Adhesion Force (vs. protein-coated tip) | 0.8 ± 0.2 nN | 2.5 ± 0.6 nN | Single-Point Force Spectroscopy |
5. Experimental Workflow and Data Integration Visualization
Title: AFM Benchmarking Workflow for 3D Biomaterials
Establishing Correlations Between AFM Surface Metrics and Biological Response (Cell Adhesion, Protein Adsorption)
Within the broader thesis on leveraging Atomic Force Microscopy (AFM) for advanced surface analysis of 3D-printed biomaterials, this application note addresses a critical translational step: linking quantitative AFM-derived surface metrics to subsequent biological performance. For tissue engineering scaffolds and implantable devices, initial protein adsorption and cellular adhesion are pivotal events dictating long-term integration and function. This document provides protocols and frameworks for establishing predictive correlations between nanoscale surface characteristics and these biological responses.
Key AFM Surface Metrics Relevant to Biological Response
The following table summarizes primary AFM-measurable parameters and their hypothesized influence on biological interactions.
Table 1: AFM Surface Metrics and Their Biological Significance
| AFM Metric | Description | Hypothesized Impact on Biological Response |
|---|---|---|
| Roughness (Ra, Rq) | Average deviation from a mean plane. | Moderate roughness (50-200 nm) often enhances protein adsorption and cell adhesion by increasing surface area and site availability. |
| Surface Skewness (Rsk) | Measure of symmetry of height distribution. | Positive Rsk (peaks) may promote focal adhesion formation; negative Rsk (valleys) might influence protein conformation. |
| Surface Kurtosis (Rku) | Measure of 'peakedness' or 'sharpness' of the surface. | High Rku (spiky peaks) could lead to localized stress concentrations affecting cell membrane integrity. |
| Nanoscale Elasticity/Modulus | Derived from force-distance curves. | Stiffer substrates (higher modulus) typically promote stronger cell adhesion and spreading via mechanotransduction. |
| Surface Adhesion Force | Measured via force spectroscopy. | Higher nanoscale adhesion can correlate with increased nonspecific protein adsorption and integrin binding. |
| Surface Texture & Orientation | Anisotropy ratio from Fourier analysis. | Directional texture can guide protein fibril alignment and direct cell migration (contact guidance). |
Research Reagent Solutions Toolkit
Table 2: Essential Materials for Integrated AFM-Biology Studies
| Item / Reagent | Function in Protocol |
|---|---|
| AFM Probes (e.g., RTESPA-300) | Silicon probes for high-resolution topography imaging in liquid. |
| CSC38 / ContGB | Cantilevers with colloidal tips for reliable force spectroscopy on soft biological samples. |
| Bovine Serum Albumin (BSA), Fluorescently Tagged | Model protein for adsorption studies; fluorescence enables quantification post-AFM. |
| Fibronectin or Vitronectin | Key adhesion proteins for studying specific integrin-mediated cell attachment. |
| Human Mesenchymal Stem Cells (hMSCs) or NIH/3T3 | Standard cell models for adhesion assays. |
| Fluorescent Phalloidin & DAPI | Stain F-actin and nuclei to visualize cell spreading and count adherent cells. |
| Serum-Free Cell Culture Medium | Used for controlled protein adsorption phases to eliminate serum variable. |
| Atomic Force Microscope with Liquid Cell | Enables imaging and force measurement under physiological conditions. |
| Confocal Microscope or Fluorescence Plate Reader | For parallel quantification of protein adsorption or cell adhesion. |
Experimental Protocols
Protocol 1: Correlating Topography with Protein Adsorption
Objective: To quantify the adsorption of a model protein (e.g., BSA) on 3D-printed material surfaces with varying AFM-measured roughness. Materials: 3D-printed polymer samples, PBS, Fluorescently tagged BSA (BSA-FITC), AFM with liquid cell, fluorescence microscope/plate reader.
Surface Characterization:
- Image at least three 50x50 µm areas per sample in PBS using tapping mode AFM.
- Calculate Ra, Rq, Rsk, and Rku using AFM software. Record nanoscale adhesion maps via force volume if possible.
Controlled Protein Adsorption:
- Incubate pre-characterized samples in BSA-FITC solution (1 mg/mL in PBS) for 1 hour at 37°C.
- Rinse samples 3x gently with PBS to remove loosely adsorbed protein.
Quantification:
- Measure fluorescence intensity (Ex/Em: 495/519 nm) per sample area using a plate reader or analyze fluorescence micrographs.
- Normalize intensity to the sample with the lowest Ra.
Correlation Analysis:
- Plot fluorescence intensity (protein adsorption) versus each AFM metric (Ra, Rsk, etc.) and perform linear or non-linear regression analysis.
Protocol 2: Correlating Nanomechanics with Cell Adhesion
Objective: To link local surface elasticity (modulus) of a 3D-printed hydrogel to initial cell adhesion density. Materials: 3D-printed hydrogel samples, serum-containing medium, serum-free medium, NIH/3T3 cells, AFM with colloidal probe, fluorescent stains, confocal microscope.
Nanomechanical Mapping:
- Perform force-volume mapping or PeakForce QNM on hydrated samples in serum-free medium.
- Derive an elasticity/modulus map and calculate the average modulus for each sample.
Cell Adhesion Assay:
- Seed cells onto characterized samples at a density of 20,000 cells/cm² in serum-containing medium.
- Allow cells to adhere for 2 hours (or a standardized time).
Adhesion Quantification:
- Fix cells, stain with phalloidin (F-actin) and DAPI (nuclei).
- Acquire 5 random confocal images per sample. Count nuclei to determine adherent cell density.
Correlation Analysis:
- Plot adherent cell density versus the average local elastic modulus for each sample.
Visualization of Workflows and Pathways
Diagram 1: Integrated AFM-Biology Correlation Workflow
Diagram 2: Surface Metrics to Cell Adhesion Signaling Path
Conclusion
AFM has emerged as an indispensable, high-resolution tool for the quantitative surface characterization of 3D-printed biomedical materials, bridging the critical gap between print parameters, nano-scale structure, and ultimate performance. By mastering foundational principles, methodological protocols, and troubleshooting strategies, researchers can reliably extract vital data on topography, roughness, and nanomechanics. Validated against complementary techniques, AFM data provides a robust framework for quality control, process optimization, and predicting biological outcomes. Future directions point toward high-speed AFM for in-situ monitoring, automated analysis for high-throughput screening of print conditions, and the establishment of standardized AFM-based metrics for regulatory approval of 3D-printed medical products. This nanoscale insight is pivotal for advancing personalized implants, tissue engineering scaffolds, and next-generation drug delivery systems from the lab bench to clinical reality.